Photoreceptor-Like Signal Transduction Between Polymer-Based Protocells
- PMID: 39491508
- PMCID: PMC11756044
- DOI: 10.1002/adma.202413981
Photoreceptor-Like Signal Transduction Between Polymer-Based Protocells
Abstract
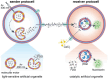
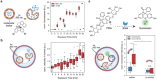
Deciphering inter- and intracellular signaling pathways is pivotal for understanding the intricate communication networks that orchestrate life's dynamics. Communication models involving bottom-up construction of protocells are emerging but often lack specialized compartments sufficiently robust and hierarchically organized to perform spatiotemporally defined signaling. Here, the modular construction of communicating polymer-based protocells designed to mimic the transduction of information in retinal photoreceptors is presented. Microfluidics is used to generate polymeric protocells subcompartmentalized by specialized artificial organelles. In one protocell population, light triggers artificial organelles with membrane-embedded photoresponsive rotary molecular motors to set off a sequence of reactions starting with the release of encapsulated signaling molecules into the lumen. Intercellular communication is mediated by signal transfer across membranes to protocells containing catalytic artificial organelles as subcompartments, whose signal conversion can be modulated by environmental calcium. Signal propagation also requires selective permeability of the diverse compartments. By segregating artificial organelles in distinct protocells, a sequential chain of reactions mediating intercellular communication is created that is further modulated by adding extracellular messengers. This connective behavior offers the potential for a deeper understanding of signaling pathways and faster integration of proto- and living cells, with the unique advantage of controlling each step by bio-relevant signals.
Keywords: artificial organelles; cell mimics; molecular motors; protocell communication and signaling.
© 2024 The Author(s). Advanced Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bradshaw R. A., Dennis E. A., Handbook of Cell Signaling, Academic Press, Cambridge, Massachusetts: 2009.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

