Construction of a Prognostic Model for Mitochondria and Macrophage Polarization Correlation in Glioma Based on Single-Cell and Transcriptome Sequencing
- PMID: 39491527
- PMCID: PMC11532235
- DOI: 10.1111/cns.70083
Construction of a Prognostic Model for Mitochondria and Macrophage Polarization Correlation in Glioma Based on Single-Cell and Transcriptome Sequencing
Abstract
Background: Numerous diseases are associated with the interplay of mitochondrial and macrophage polarization. However, the correlation of mitochondria-related genes (MRGs) and macrophage polarization-related genes (MPRGs) with the prognosis of glioma remains unclear. This study aimed to examine this relationship based on bioinformatic analysis.
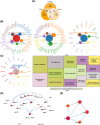
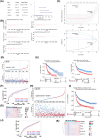
Methods: Glioma-related datasets (TCGA-GBMLGG, mRNA-seq-325, mRNA-seq-693, GSE16011, GSE4290, and GSE138794) were included in this study. The intersection genes were obtained by overlapping differentially expressed genes (DEGs) from differential expression analysis in GSE16011, key module genes from WGCNA, and MRGs. Subsequently, the intersection genes were further screened to obtain prognostic genes. Following this, a risk model was developed and verified. After that, independent prognostic factors were identified, followed by the construction of a nomogram and subsequent evaluation of its predictive ability. Furthermore, immune microenvironment analysis and expression validation were implemented. The GSE138794 dataset was utilized to evaluate the expression of prognostic genes at a cellular level, followed by conducting an analysis on cell-to-cell communication. Finally, the results were validated in different datasets and tissue samples from patients.
Results: ECI2, MCCC2, OXCT1, SUCLG2, and CPT2 were identified as prognostic genes for glioma. The risk model constructed based on these genes in TCGA-GBMLGG demonstrated certain accuracy in predicting the occurrence of glioma. Additionally, the nomogram constructed based on risk score and grade exhibited strong performance in predicting patient survival. Significant differences were observed in the proportion of 27 immune cell types (e.g., activated B cells and macrophages) and the expression of 32 immune checkpoints (e.g., CD70, CD200, and CD48) between the two risk groups. Single-cell RNA sequencing showed that CPT2, ECI2, and SUCLG2 were highly expressed in oligodendrocytes, neural progenitor cells, and BMDMs, respectively. The results of cell-cell communication analysis revealed that both oligodendrocytes and BMDMs exhibited a substantial number of interactions with high strength.
Conclusion: This study revealed five genes associated with the prognosis of glioma (ECI2, MCCC2, OXCT1, SUCLG2, and CPT2), providing novel insights into individualized treatment and prognosis.
Keywords: glioma; immune microenvironment; macrophage polarization; mitochondria; risk model; single‐cell RNA‐seq analysis.
© 2024 The Author(s). CNS Neuroscience & Therapeutics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Xu S., Tang L., Li X., Fan F., and Liu Z., “Immunotherapy for Glioma: Current Management and Future Application,” Cancer Letters 476 (2020): 1–12. - PubMed
-
- Anderson N. R., Minutolo N. G., Gill S., and Klichinsky M., “Macrophage‐Based Approaches for Cancer Immunotherapy,” Cancer Research 81, no. 5 (2021): 1201–1208. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

