The Complement Factor H (Y402H) risk polymorphism for age-related macular degeneration affects metabolism and response to oxidative stress in the retinal pigment epithelium
- PMID: 39491736
- PMCID: PMC11662989
- DOI: 10.1016/j.freeradbiomed.2024.10.307
The Complement Factor H (Y402H) risk polymorphism for age-related macular degeneration affects metabolism and response to oxidative stress in the retinal pigment epithelium
Abstract
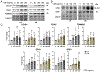
Age-related macular degeneration (AMD), the leading cause of central vision loss in the elderly, involves death of the retinal pigment epithelium (RPE) and light-sensing photoreceptors. This multifactorial disease includes contributions from both genetic and environmental risk factors. The current study examined the effect of the Y402H polymorphism of Complement Factor H (CFH, rs1061170) and cigarette smoke, predominant genetic and environmental risk factors associated with AMD. We used targeted and discovery-based approaches to identify genotype-dependent responses to chronic oxidative stress induced by cigarette smoke extract (CSE) in RPE differentiated from induced pluripotent stem cells (iPSC) derived from human donors harboring either the low risk (LR) or high risk (HR) CFH genotype. Chronic CSE altered the metabolic profile in both LR and HR iPSC-RPE and caused a dose-dependent reduction in mitochondrial function despite an increase in mitochondrial content. Notably, cells with the HR CFH SNP showed a greater reduction in maximal respiration and ATP production. Significant genotype-dependent changes in the proteome were observed for HR RPE at baseline (cytoskeleton, MAPK signaling) and after CSE exposure, where a less robust upregulation of the antioxidants and significant downregulation in proteins involved in nucleic acid metabolism and membrane trafficking were noted compared to LR cells. In LR cells, uniquely upregulated proteins were involved in lipid metabolism and chemical detoxification. These genotype-dependent differences at baseline and in response to chronic CSE exposure suggest a broader role for CFH in modulating the response to oxidative stress in RPE and provides insight into the interaction between environmental and genetic factors in AMD pathogenesis.
Keywords: Bioinformatics; Cigarette smoke extract; Metabolomics; Proteomics; iPSC-RPE.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest None.
Figures


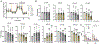


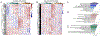

References
-
- Friedman DS, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol, 2004. 122(4): p. 564–72. - PubMed
-
- Wong WL, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health, 2014. 2(2): p. e106–16. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

