Causal effects of gut microbiota on gout and hyperuricemia: insights from genome-wide Mendelian randomization, RNA-sequencing, 16S rRNA sequencing, and metabolomes
- PMID: 39492788
- PMCID: PMC11598824
- DOI: 10.1042/BSR20240595
Causal effects of gut microbiota on gout and hyperuricemia: insights from genome-wide Mendelian randomization, RNA-sequencing, 16S rRNA sequencing, and metabolomes
Abstract
Background: This study investigated the causal relationship between gut microbiota (GM), serum metabolome, and host transcriptome in the development of gout and hyperuricemia (HUA) using genome-wide association studies (GWAS) data and HUA mouse model experiments.
Methods: Mendelian randomization (MR) analysis of GWAS summary statistics was performed using an inverse variance weighted (IVW) approach to determine or predict the causal role of the GM on gout. The HUA mouse model was used to characterize changes in the gut microbiome, host metabolome, and host kidney transcriptome by integrating cecal 16S rRNA sequencing, untargeted serum metabolomics, and host mRNA sequencing.
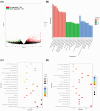
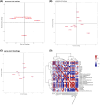
Results: Our analysis demonstrated causal effects of seven GM taxa on gout, including genera of Ruminococcus, Odoribacter, and Bacteroides. Thirty eight immune cell traits were associated with gout. Dysbiosis of Dubosiella, Lactobacillus, Bacteroides, Alloprevotella, and Lachnospiraceae_NK4A136_group genera were associated with changes in the serum metabolites and kidney transcriptome of the HUA model mice. The changes in the gut microbiome of the HUA model mice correlated significantly with alterations in the levels of serum metabolites such as taurodeoxycholic acid, phenylacetylglycine, vanylglycol, methyl hexadecanoic acid, carnosol, 6-aminopenicillanic acid, sphinganine, p-hydroxyphenylacetic acid, pyridoxamine, and de-o-methylsterigmatocystin, and expression of kidney genes such as CNDP2, SELENOP, TTR, CAR3, SLC12A3, SCD1, PIGR, CD74, MFSD4B5, and NAPSA.
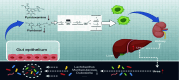
Conclusion: Our study demonstrated a causal relationship between GM, immune cells, and gout. HUA development involved alterations in the vitamin B6 metabolism because of GM dysbiosis that resulted in altered pyridoxamine and pyridoxal levels, dysregulated sphingolipid metabolism, and excessive inflammation.
Keywords: Gout; Hyperuricemia; Mendelian randomization; gut microbiota; multi-omics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


References
-
- Peng A., Lin L., Zhao M. and Sun B. (2019) Identifying mechanisms underlying the amelioration effect of Chrysanthemum morifolium Ramat. ‘Boju’ extract on hyperuricemia using biochemical characterization and UPLC-ESI-QTOF/MS-based metabolomics. Food Funct. 10, 8042–8055 10.1039/C9FO01821B - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

