Targeting the DNA damage response in cancer
- PMID: 39492835
- PMCID: PMC11527828
- DOI: 10.1002/mco2.788
Targeting the DNA damage response in cancer
Abstract
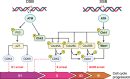
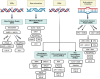
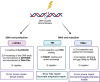
DNA damage response (DDR) pathway is the coordinated cellular network dealing with the identification, signaling, and repair of DNA damage. It tightly regulates cell cycle progression and promotes DNA repair to minimize DNA damage to daughter cells. Key proteins involved in DDR are frequently mutated/inactivated in human cancers and promote genomic instability, a recognized hallmark of cancer. Besides being an intrinsic property of tumors, DDR also represents a unique therapeutic opportunity. Indeed, inhibition of DDR is expected to delay repair, causing persistent unrepaired breaks, to interfere with cell cycle progression, and to sensitize cancer cells to several DNA-damaging agents, such as radiotherapy and chemotherapy. In addition, DDR defects in cancer cells have been shown to render these cells more dependent on the remaining pathways, which could be targeted very specifically (synthetic lethal approach). Research over the past two decades has led to the synthesis and testing of hundreds of small inhibitors against key DDR proteins, some of which have shown antitumor activity in human cancers. In parallel, the search for synthetic lethality interaction is broadening the use of DDR inhibitors. In this review, we discuss the state-of-art of ataxia-telangiectasia mutated, ataxia-telangiectasia-and-Rad3-related protein, checkpoint kinase 1, Wee1 and Polθ inhibitors, highlighting the results obtained in the ongoing clinical trials both in monotherapy and in combination with chemotherapy and radiotherapy.
Keywords: ATM; ATR; Chk1; DNA damage response; Polθ; Wee1; solid tumors.
© 2024 The Author(s). MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures
References
-
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double‐strand break repair pathway choice. Mol Cell. 2012;47(4):497‐510. - PubMed
-
- Groelly FJ, Fawkes M, Dagg RA, Blackford AN, Tarsounas M. Targeting DNA damage response pathways in cancer. Nat Rev Cancer. 2023;23(2):78‐94. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646‐674. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
