Genomic and phenotypic characterization of a Clostridioides difficile strain of the epidemic ST37 type from China
- PMID: 39492989
- PMCID: PMC11527712
- DOI: 10.3389/fcimb.2024.1412408
Genomic and phenotypic characterization of a Clostridioides difficile strain of the epidemic ST37 type from China
Abstract
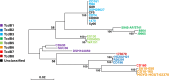
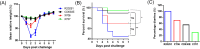
Clostridioides difficile strains of sequence type (ST) 37, primarily including PCR ribotype (RT) 017, are prevalent in mainland China. Our study aimed to compare the major virulence factors of an epidemic C. difficile isolate of ST37 type (Xy06) from China with the well-characterized C. difficile reference strains R20291 (RT027) and CD630E (ST54), as well as a Chinese ST54 strain (Xy07) isolated from the same hospital. The Xy06 genome was predicted to harbor two complete prophages and several transposon-like elements. Comparative analysis of PaLoc revealed a truncated tcdA gene, a functional tcdB gene, a functional tcdC gene, and well-conserved tcdR and tcdE genes. Phenotypic comparisons showed that Xy06 was a robust producer of TcdB, readily sporulated and germinated, and strongly bound to human gut epithelial cells. In a mouse model of C. difficile infection, Xy06 was more virulent than strains CD630E and Xy07 and was comparable to strain R20291 in virulence. Our data suggest the potential threat of the epidemic ST37 strains in China.
Keywords: Clostridioides difficile; RT017; ST37; pathogenicity locus (PaLoc); toxin; virulence.
Copyright © 2024 Li, Heuler, Zhu, Meng, Chakraborty, Harmanus, Wang, Peng, Smits, Wu and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

