Proteomic profiling of neutrophils and plasma in community-acquired pneumonia reveals crucial proteins in diverse biological pathways linked to clinical outcome
- PMID: 39493755
- PMCID: PMC11527607
- DOI: 10.3389/fimmu.2024.1470383
Proteomic profiling of neutrophils and plasma in community-acquired pneumonia reveals crucial proteins in diverse biological pathways linked to clinical outcome
Abstract
Introduction: Neutrophils play a dichotomous role in community-acquired pneumonia (CAP), providing protection and potentially causing damage. Existing research on neutrophil function in CAP relies on animal studies, leaving a gap in patient-centered investigations.
Methods: We used mass spectrometry to characterize the neutrophil proteome of moderately ill CAP patients at general ward admission and related the proteome to controls and clinical outcomes.
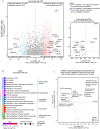
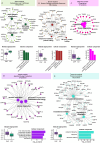
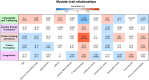
Results: We prospectively included 57 CAP patients and 26 controls and quantified 3482 proteins in neutrophil lysates and 386 proteins in concurrently collected plasma. The extensively studied granule-related proteins in animal models did not drive the neutrophil proteome changes associated with human CAP. Proteome alterations were primarily characterized by an increased abundance of proteins related to (aerobic) metabolic activity and (m)RNA translation/processing, concurrent with a diminished presence of cytoskeletal organization-related proteins (all pathways p<0.001). Higher and lower abundances of specific proteins, primarily constituents of these pathways, were associated with prolonged time to clinical stability in CAP. Moreover, we identified a pronounced presence of platelet-related proteins in neutrophil lysates of particularly viral CAP patients, suggesting the existence of neutrophil-platelet complexes in non-critically ill CAP patients. Of the proteins measured in neutrophils, 4.3% were detected in plasma.
Discussion: Our study presents new perspectives on the neutrophil proteome associated with CAP, laying the groundwork for forthcoming patient-centred investigations. Our results could pave the way for targeted strategies to fine-tune neutrophil responses, potentially improving CAP outcomes.
Keywords: community-acquired pneumonia; general ward; innate immunity; mass spectrometry; neutrophil; pneumonia; proteomics.
Copyright © 2024 Michels, Chouchane, de Brabander, de Vos, Faber, Douma, Smit, Wiersinga, Biggelaar, Poll and Hoogendijk.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

