Human SKI component SKIV2L regulates telomeric DNA-RNA hybrids and prevents telomere fragility
- PMID: 39493885
- PMCID: PMC11530851
- DOI: 10.1016/j.isci.2024.111096
Human SKI component SKIV2L regulates telomeric DNA-RNA hybrids and prevents telomere fragility
Abstract
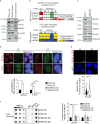
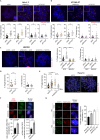
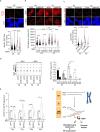
Super killer (SKI) complex is a well-known cytoplasmic 3'-5' mRNA decay complex that functions with the exosome to degrade excessive and aberrant mRNAs, is implicated with the extraction of mRNA at stalled ribosomes, tackling aberrant translation. Here, we show that SKIV2L and TTC37 of the hSKI complex are present within the nucleus, localize on chromatin and at some telomeres during the G2 cell cycle phase. In cells, SKIV2L prevents telomere replication stress, independently of its helicase domain, and increases the stability of telomere DNA-RNA hybrids in G2. We further demonstrate that purified hSKI complex binds telomeric DNA and RNA substrates in vitro and SKIV2L association with telomeres is dependent on DNA-RNA hybrids. Taken together, our results provide a nuclear function for SKIV2L of the hSKI complex in overcoming replication stress at telomeres mediated by its recruitment to DNA-RNA hybrid structures in G2 and thus maintaining telomere stability.
Keywords: Cell biology; Molecular biology.
© 2024 Imperial College London.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


References
LinkOut - more resources
Full Text Sources

