The genome sequence of Inga leiocalycina Benth
- PMID: 39494196
- PMCID: PMC11531642
- DOI: 10.12688/wellcomeopenres.23131.1
The genome sequence of Inga leiocalycina Benth
Abstract
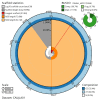
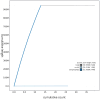
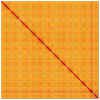
We present a genome assembly from an individual of Inga leiocalycina (Streptophyta; Magnoliopsida; Fabales; Fabaceae). The genome sequence has a total length of 948.00 megabases. Most of the assembly is scaffolded into 13 chromosomal pseudomolecules. The assembled mitochondrial genome sequences have lengths of 1,019.42 and 98.74 kilobases, and the plastid genome assembly is 175.51 kb long. Gene annotation of the nuclear genome assembly on Ensembl identified 33,457 protein-coding genes.
Keywords: Fabales; Inga leiocalycina; chromosomal; genome sequence.
Copyright: © 2024 Schley RJ et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
-
- Bates A, Clayton-Lucey I, Howard C: Sanger Tree of Life HMW DNA fragmentation: diagenode Megaruptor ®3 for LI PacBio. protocols.io. 2023. 10.17504/protocols.io.81wgbxzq3lpk/v1 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

