Forkhead box D subfamily genes in colorectal cancer: potential biomarkers and therapeutic targets
- PMID: 39494294
- PMCID: PMC11529599
- DOI: 10.7717/peerj.18406
Forkhead box D subfamily genes in colorectal cancer: potential biomarkers and therapeutic targets
Abstract
Background: The forkhead box (FOX) family members regulate gene transcription and expression. FOX family members regulate various biological processes, such as cell proliferation and tumorigenesis. FOXD, a FOX protein subfamily, is associated with poor prognosis for various cancers. However, the potential clinical value of FOXD subfamily members in colorectal cancer (CRC) has not yet been elucidated. Therefore, in this study, we aimed to determine the role of the FOXD subfamily members in CRC development.
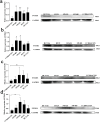
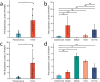
Methods: Using HTSeq-count data, clinical data, and single-nucleotide polymorphisms (obtained from The Cancer Genome Atlas Project), and bioinformatics analyses (using DESEQ2 software), we identified differentially expressed genes (DEGs) in CRC. Next, each DEG expression was validated in vitro using reverse transcription-quantitative polymerase chain reaction, western blotting, and immunohistochemistry (IHC).
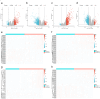
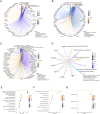
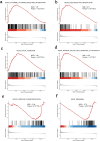
Results: Among the FOXD subfamily members, the area under the receiver operating characteristic curve of FOXD3 was 0.949, indicating that FOXD3 has a high overall diagnostic accuracy for CRC. Gene Set Enrichment Analysis revealed that FOXD-DEGs were mainly related to pathways such as cytokine, cytokine, and extracellular matrix receptor interactions. Kaplan-Meier curves and nomograms showed that FOXD1, FOXD3, and FOXD4 were prognostically significant. In conclusion, FOXD subfamily members (especially FOXD3) could serve as diagnostic and prognostic biomarkers for CRC and an immunotherapy target in patients with CRC.
Keywords: Colorectal cancer; Forkhead box D subfamily; Immune cells; Prognosis.
© 2024 Chen et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Adeleke S, Haslam A, Choy A, Diaz-Cano S, Galante JR, Mikropoulos C, Boussios S. Microsatellite instability testing in colorectal patients with Lynch syndrome: lessons learned from a case report and how to avoid such pitfalls. Personalized Medicine. 2022;19(4):277–286. doi: 10.2217/pme-2021-0128. - DOI - PubMed
-
- Bademci G, Abad C, Incesulu A, Elian F, Reyahi A, Diaz-Horta O, Cengiz FB, Sineni CJ, Seyhan S, Atli EI, Basmak H, Demir S, Nik AM, Footz T, Guo S, Duman D, Fitoz S, Gurkan H, Blanton SH, Walter MA, Carlsson P, Walz K, Tekin M. FOXF2 is required for cochlear development in humans and mice. Human Molecular Genetics. 2019;28(8):1286–1297. doi: 10.1093/hmg/ddy431. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

