Exploring the therapeutic mechanisms of millet in obesity through molecular docking, pharmacokinetics, and dynamic simulation
- PMID: 39494311
- PMCID: PMC11528469
- DOI: 10.3389/fnut.2024.1453819
Exploring the therapeutic mechanisms of millet in obesity through molecular docking, pharmacokinetics, and dynamic simulation
Abstract
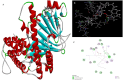
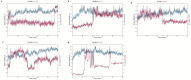
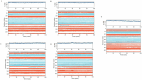
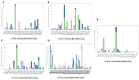
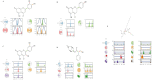
Obesity, a prevalent global health concern, is characterized by excessive fat accumulation, which confers significant nutritional and health risks, including a shortened lifespan and diminished wellbeing. Central to the regulation of energy balance and food intake is the fat mass and obesity-associated (FTO) protein, which modulates the interplay between caloric consumption and energy expenditure. Given its pivotal role in obesity regulation, the identification of effective inhibitors targeting the FTO protein is imperative for developing therapeutic interventions. Currently available anti-obesity drugs are often plagued by undesirable side effects. In contrast, natural plant-derived bioactive compounds are gaining prominence in the pharmaceutical industry due to their efficacy and lower incidence of adverse effects. Little Millet, a traditional cereal known for its rich nutritional profile and high satiety index, was investigated in this study using molecular docking and dynamics simulation approach for its potential as an anti-obesity agent. Our research demonstrates that four bioactive compounds from Little Millet exhibit superior binding energies ranging from 7.22 to 8.83 kcal/mol, compared to the standard anti-obesity drug, orlistat, which has a binding energy of 5.96 kcal/mol. These compounds fulfilled all drug-like criteria, including the Lipinski, Ghose, Veber, Egan, and Muegge rules, and exhibited favorable profiles in terms of distribution, metabolism, and prolonged half-life without toxicity. Conversely, orlistat was associated with hepatotoxicity, a reduced half-life, and multiple violations of drug-likeness parameters, undermining its efficacy. Molecular dynamics simulations and Gibbs free energy assessments revealed that the four identified compounds maintain stable interactions with key residues in the FTO protein's active site. We propose further validation through extensive In vitro, In vivo, and clinical studies to ascertain the therapeutic potential of these compounds in combating obesity.
Keywords: ADMET; binding free energy; human fat mass and obesity associated protein (FTO); little millet; molecular docking and molecular dynamics (MD) simulation; obesity.
Copyright © 2024 Lakhani, Hamid, Gupta, Prajapati, Prabha, Patel and Suthar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
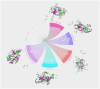
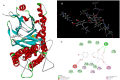
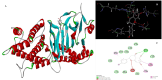
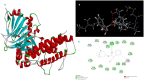
Figures



References
LinkOut - more resources
Full Text Sources

