Longitudinal genomic surveillance of a UK intensive care unit shows a lack of patient colonisation by multi-drug-resistant Gram-negative bacterial pathogens
- PMID: 39494554
- PMCID: PMC11533117
- DOI: 10.1099/mgen.0.001314
Longitudinal genomic surveillance of a UK intensive care unit shows a lack of patient colonisation by multi-drug-resistant Gram-negative bacterial pathogens
Abstract
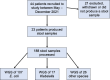
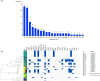
Vulnerable patients in an intensive care unit (ICU) setting are at high risk of infection from bacteria including gut-colonising Escherichia coli and Klebsiella species. Complex ICU procedures often depend on successful antimicrobial treatment, underscoring the importance of understanding the extent of patient colonisation by multi-drug-resistant organisms (MDROs) in large UK ICUs. Previous work on ICUs globally uncovered high rates of colonisation by transmission of MDROs, but the situation in UK ICUs is less understood. Here, we investigated the diversity and antibiotic resistance gene (ARG) carriage of bacteria present in one of the largest UK ICUs at the Queen Elizabeth Hospital Birmingham (QEHB), focusing primarily on E. coli as both a widespread commensal and a globally disseminated multi-drug-resistant pathogen. Samples were taken during highly restrictive coronavirus disease 2019 (COVID-19) control measures from May to December 2021. Whole-genome and metagenomic sequencing were used to detect and report strain-level colonisation of patients, focusing on E. coli sequence types (STs), their colonisation dynamics and antimicrobial resistance gene carriage. We found a lack of multi-drug resistance (MDR) in the QEHB. Only one carbapenemase-producing organism was isolated, a Citrobacter carrying bla KPC-2. There was no evidence supporting the spread of this strain, and there was little evidence overall of nosocomial acquisition or circulation of colonising E. coli. Whilst 22 different E. coli STs were identified, only 1 strain of the pandemic ST131 lineage was isolated. This ST131 strain was non-MDR and was found to be a clade A strain, associated with low levels of antibiotic resistance. Overall, the QEHB ICU had very low levels of pandemic or MDR strains, a result that may be influenced in part by the strict COVID-19 control measures in place at the time. Employing some of these infection prevention and control measures where reasonable in all ICUs might therefore assist in maintaining low levels of nosocomial MDR.
Keywords: AMR; E. coli; ICU.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- O’Neill J. The review on antimicrobial resistance - antimicrobial resistance:tackling a crisis for the health and wealth of nations. 2014.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

