A genome-engineered tool set for Drosophila TGF-β/BMP signaling studies
- PMID: 39494616
- PMCID: PMC11607693
- DOI: 10.1242/dev.204222
A genome-engineered tool set for Drosophila TGF-β/BMP signaling studies
Abstract
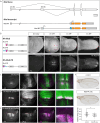
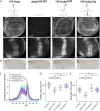
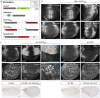
Ligands of the TGF-β/BMP superfamily are crucially involved in the regulation of growth, patterning and organogenesis and can act as long-range morphogens. Essential for understanding TGF-β/BMP signaling dynamics and regulation are tools that allow monitoring and manipulating pathway components at physiological expression levels and endogenous spatiotemporal patterns. We used genome engineering to generate a comprehensive library of endogenously epitope- or fluorescent-tagged versions of receptors, co-receptors, transcription factors and key feedback regulators of the Drosophila BMP and Activin signaling pathways. We demonstrate that the generated alleles are biologically active and can be used for assessing tissue and subcellular distribution of the corresponding proteins. Furthermore, we show that the genomic platforms can be used for in locus structure-function and cis-regulatory analyses. Finally, we present a complementary set of protein binder-based tools, which allow visualization as well as manipulation of the stability and subcellular localization of epitope-tagged proteins, providing new tools for the analysis of BMP signaling and beyond.
Keywords: Drosophila development; Endogenous protein tagging; Functionalized protein binder tools; Oogenesis; TGF-β/BMP signaling; Wing imaginal disc.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

