Insights into the regulatory mechanisms and application prospects of the transcription factor Cra
- PMID: 39494897
- PMCID: PMC11577769
- DOI: 10.1128/aem.01228-24
Insights into the regulatory mechanisms and application prospects of the transcription factor Cra
Erratum in
-
Erratum for Huang et al., "Insights into the regulatory mechanisms and application prospects of the transcription factor Cra".Appl Environ Microbiol. 2025 Feb 19;91(2):e0004725. doi: 10.1128/aem.00047-25. Epub 2025 Jan 31. Appl Environ Microbiol. 2025. PMID: 39887240 Free PMC article. No abstract available.
Abstract
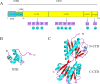
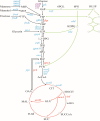
Cra (catabolite repressor/activator) is a global transcription factor (TF) that plays a pleiotropic role in controlling the transcription of several genes involved in carbon utilization and energy metabolism. Multiple studies have investigated the regulatory mechanism of Cra and its rational use for metabolic regulation, but due to the complexity of its regulation, there remain challenges in the efficient use of Cra. Here, the structure, mechanism of action, and regulatory function of Cra in carbon and nitrogen flow are reviewed. In addition, this paper highlights the application of Cra in metabolic engineering, including the promotion of metabolite biosynthesis, the regulation of stress tolerance and virulence, the use of a Cra-based biosensor, and its coupling with other transcription factors. Finally, the prospects of Cra-related regulatory strategies are discussed. This review provides guidance for the rational design and construction of Cra-based metabolic regulation systems.
Keywords: carbon metabolism; metabolic regulation; transcription factor Cra.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Ramseier TM, Nègre D, Cortay JC, Scarabel M, Cozzone AJ, Saier MH Jr. 1993. In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J Mol Biol 234:28–44. doi: 10.1006/jmbi.1993.1561 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

