A Target Class Ligandability Evaluation of WD40 Repeat-Containing Proteins
- PMID: 39495097
- PMCID: PMC11770632
- DOI: 10.1021/acs.jmedchem.4c02010
A Target Class Ligandability Evaluation of WD40 Repeat-Containing Proteins
Abstract
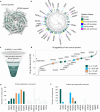
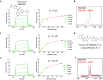
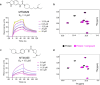
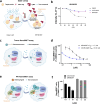
Target class-focused drug discovery has a strong track record in pharmaceutical research, yet public domain data indicate that many members of protein families remain unliganded. Here we present a systematic approach to scale up the discovery and characterization of small molecule ligands for the WD40 repeat (WDR) protein family. We developed a comprehensive suite of protocols for protein production, crystallography, and biophysical, biochemical, and cellular assays. A pilot hit-finding campaign using DNA-encoded chemical library selection followed by machine learning (DEL-ML) to predict ligands from virtual libraries yielded first-in-class, drug-like ligands for 7 of the 16 WDR domains screened, thus demonstrating the broader ligandability of WDRs. This study establishes a template for evaluation of protein family wide ligandability and provides an extensive resource of WDR protein biochemical and chemical tools, knowledge, and protocols to discover potential therapeutics for this highly disease-relevant, but underexplored target class.
Conflict of interest statement
The authors declare the following competing financial interest(s): BLS, BG, JSD, JZ, JWC, MvR, PR, SK, and TK are current or past employees and shareholders of Relay Therapeutics.
Figures



Comment in
-
A Systematic Blueprint to Ligand the Proteome.J Med Chem. 2025 Jan 23;68(2):1090-1091. doi: 10.1021/acs.jmedchem.4c03091. Epub 2025 Jan 6. J Med Chem. 2025. PMID: 39761356
References
-
- Yeh J. I.; Levine A. S.; Du S.; Chinte U.; Ghodke H.; Wang H.; Shi H.; Hsieh C. L.; Conway J. F.; Van Houten B.; Rapic-Otrin V. Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2012, 109 (41), E2737-4610.1073/pnas.1110067109. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

