TMEM16 and OSCA/TMEM63 proteins share a conserved potential to permeate ions and phospholipids
- PMID: 39495104
- PMCID: PMC11534332
- DOI: 10.7554/eLife.96957
TMEM16 and OSCA/TMEM63 proteins share a conserved potential to permeate ions and phospholipids
Abstract
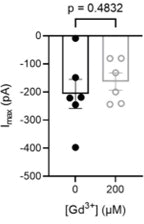
The calcium-activated TMEM16 proteins and the mechanosensitive/osmolarity-activated OSCA/TMEM63 proteins belong to the Transmembrane Channel/Scramblase (TCS) superfamily. Within the superfamily, OSCA/TMEM63 proteins, as well as TMEM16A and TMEM16B, are thought to function solely as ion channels. However, most TMEM16 members, including TMEM16F, maintain an additional function as scramblases, rapidly exchanging phospholipids between leaflets of the membrane. Although recent studies have advanced our understanding of TCS structure-function relationships, the molecular determinants of TCS ion and lipid permeation remain unclear. Here, we show that single mutations along the transmembrane helix (TM) 4/6 interface allow non-scrambling TCS members to permeate phospholipids. In particular, this study highlights the key role of TM 4 in controlling TCS ion and lipid permeation and offers novel insights into the evolution of the TCS superfamily, suggesting that, like TMEM16s, the OSCA/TMEM63 family maintains a conserved potential to permeate ions and phospholipids.
Keywords: OSCA; TMEM16; TMEM63; evolution; human; ion channel; molecular biophysics; scramblase; structural biology.
© 2024, Lowry, Liang et al.
Conflict of interest statement
AL, PL, MS, YW, ZP, HY, YZ No competing interests declared
Figures



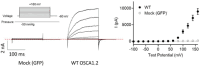





Update of
-
TMEM16 and OSCA/TMEM63 proteins share a conserved potential to permeate ions and phospholipids.bioRxiv [Preprint]. 2024 Sep 3:2024.02.04.578431. doi: 10.1101/2024.02.04.578431. bioRxiv. 2024. Update in: Elife. 2024 Nov 04;13:RP96957. doi: 10.7554/eLife.96957. PMID: 38370744 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

