FOXR2 Targets LHX6+/DLX+ Neural Lineages to Drive Central Nervous System Neuroblastoma
- PMID: 39495206
- PMCID: PMC11733536
- DOI: 10.1158/0008-5472.CAN-24-2248
FOXR2 Targets LHX6+/DLX+ Neural Lineages to Drive Central Nervous System Neuroblastoma
Abstract
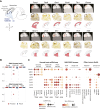
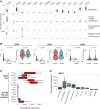
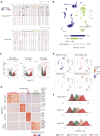
Central nervous system neuroblastoma with forkhead box R2 (FOXR2) activation (NB-FOXR2) is a high-grade tumor of the brain hemispheres and a newly identified molecular entity. Tumors express dual neuronal and glial markers, leading to frequent misdiagnoses, and limited information exists on the role of FOXR2 in their genesis. To identify their cellular origins, we profiled the transcriptomes of NB-FOXR2 tumors at the bulk and single-cell levels and integrated these profiles with large single-cell references of the normal brain. NB-FOXR2 tumors mapped to LHX6+/DLX+ lineages derived from the medial ganglionic eminence, a progenitor domain in the ventral telencephalon. In vivo prenatal Foxr2 targeting to the ganglionic eminences in mice induced postnatal cortical tumors recapitulating human NB-FOXR2-specific molecular signatures. Profiling of FOXR2 binding on chromatin in murine models revealed an association with ETS transcriptional networks, as well as direct binding of FOXR2 at key transcription factors that coordinate initiation of gliogenesis. These data indicate that NB-FOXR2 tumors originate from LHX6+/DLX+ interneuron lineages, a lineage of origin distinct from that of other FOXR2-driven brain tumors, highlight the susceptibility of ventral telencephalon-derived interneurons to FOXR2-driven oncogenesis, and suggest that FOXR2-induced activation of glial programs may explain the mixed neuronal and oligodendroglial features in these tumors. More broadly, this work underscores systematic profiling of brain development as an efficient approach to orient oncogenic targeting for in vivo modeling, critical for the study of rare tumors and development of therapeutics. Significance: Profiling the developing brain enabled rationally guided modeling of FOXR2-activated CNS neuroblastoma, providing a strategy to overcome the heterogeneous origins of pediatric brain tumors that hamper tumor modeling and therapy development. See related commentary by Orr, p. 195.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
P. Bandopadhayay reports grants from the Novartis Institute of Biomedical Research and personal fees from Day One Biopharmaceuticals outside the submitted work. No disclosures were reported by the other authors.
Figures




References
MeSH terms
Substances
Grants and funding
- R01 NS116361/NS/NINDS NIH HHS/United States
- RGPIN-2016-04911/Natural Sciences and Engineering Research Council of Canada (NSERC)
- Fonds de Recherche du Québec - Santé (FRQS)
- Genome Canada (GC)
- PJT-156086/Canadian Institutes of Health Research (CIHR)
- PJT-190271/Canadian Institutes of Health Research (CIHR)
- Fondation Charles-Bruneau (Charles Bruneau Foundation)
- Faculty of Medicine, McGill University (McGill Faculty of Medicine)
- Award RCCCEA-May22\100003/Cancer Research UK (CRUK)
- St. Baldrick's Foundation (SBF)
- Helen Gurley Brown Presidential Initiative
- V4020/Great Ormond Street Hospital Charity (GOSH)
- Rally Foundation (Rally Foundation, Inc.)
- R01NS116361/National Institute of Neurological Disorders and Stroke (NINDS)
- Cancer Research UK Cambridge Institute, University of Cambridge (CRUK CI)
- Alex's Lemonade Stand Foundation for Childhood Cancer (ALSF)
- Pedals for Pediatrics
- Lady Davis Institute for Medical Research (LDI)
- Calcul Quebec
- Kids Join the Fight
- Canadian Institutes of Health Research (CIHR)
- Griffin's Guardians
- Emily Parsons Donation
- 202021-28/Brain Research UK (BRUK)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

