Germline copy number variants and endometrial cancer risk
- PMID: 39495297
- PMCID: PMC11576655
- DOI: 10.1007/s00439-024-02707-9
Germline copy number variants and endometrial cancer risk
Abstract
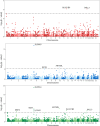
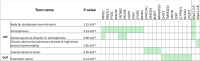
Known risk loci for endometrial cancer explain approximately one third of familial endometrial cancer. However, the association of germline copy number variants (CNVs) with endometrial cancer risk remains relatively unknown. We conducted a genome-wide analysis of rare CNVs overlapping gene regions in 4115 endometrial cancer cases and 17,818 controls to identify functionally relevant variants associated with disease. We identified a 1.22-fold greater number of CNVs in DNA samples from cases compared to DNA samples from controls (p = 4.4 × 10-63). Under three models of putative CNV impact (deletion, duplication, and loss of function), genome-wide association studies identified 141 candidate gene loci associated (p < 0.01) with endometrial cancer risk. Pathway analysis of the candidate loci revealed an enrichment of genes involved in the 16p11.2 proximal deletion syndrome, driven by a large recurrent deletion (chr16:29,595,483-30,159,693) identified in 0.15% of endometrial cancer cases and 0.02% of control participants. Together, these data provide evidence that rare copy number variants have a role in endometrial cancer susceptibility and that the proximal 16p11.2 BP4-BP5 region contains 25 candidate risk gene(s) that warrant further analysis to better understand their role in human disease.
© 2024. The Author(s).
Conflict of interest statement
Figures


References
-
- Abel HJ, Larson DE, Regier AA, Chiang C, Das I, Kanchi KL, Layer RM, Neale BM, Salerno WJ, Reeves C, Buyske S, Abecasis GR, Appelbaum E, Baker J, Banks E, Bernier RA, Bloom T, Boehnke M, Boerwinkle E, Hall IM (2020) Mapping and characterization of structural variation in 17,795 human genomes. Nature. 10.1038/s41586-020-2371-0 - PMC - PubMed
-
- Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, Casey G, Hunter DJ, Sellers TA, Gruber SB, Dunning AM, Michailidou K, Fachal L, Doheny K, Spurdle AB, Li Y, Xiao X, Romm J, Pugh E, Easton DF (2017) The oncoarray consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol Biomark Prev. 10.1158/1055-9965.EPI-16-0106 - PMC - PubMed
-
- Aune D, Navarro Rosenblatt DA, Chan DSM, Vingeliene S, Abar L, Vieira AR, Greenwood DC, Bandera EV, Norat T (2015) Anthropometric factors and endometrial cancer risk: A systematic review and dose-response meta-analysis of prospective studies. Ann Oncol. 10.1093/annonc/mdv142 - PubMed
MeSH terms
Grants and funding
- APP177524/National Health and Medical Research Council
- R01 CA128931/CA/NCI NIH HHS/United States
- R01 CA176785/CA/NCI NIH HHS/United States
- UM1 CA164920/CA/NCI NIH HHS/United States
- APP1111246/National Health and Medical Research Council
- R01 CA097396/CA/NCI NIH HHS/United States
- R01 CA058598/CA/NCI NIH HHS/United States
- P50 CA116201/CA/NCI NIH HHS/United States
- R01 CA149429/CA/NCI NIH HHS/United States
- R01 CA192393/CA/NCI NIH HHS/United States
- R01 CA116167/CA/NCI NIH HHS/United States
- R01 CA177150/CA/NCI NIH HHS/United States
- #19/460/Health Research Council of New Zealand
- R01 CA122443/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- R35 CA253187/CA/NCI NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- U19 CA148065/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA140286/CA/NCI NIH HHS/United States
- P50 CA136393/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources

