Establishment and drug resistance characterization of paired organoids using human primary colorectal cancer and matched tumor deposit specimens
- PMID: 39495391
- PMCID: PMC11534897
- DOI: 10.1007/s13577-024-01139-x
Establishment and drug resistance characterization of paired organoids using human primary colorectal cancer and matched tumor deposit specimens
Abstract
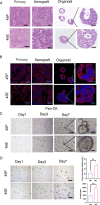
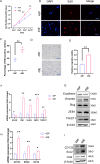
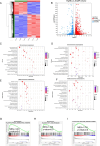
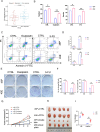
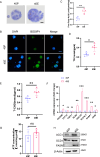
Tumor deposits (TDs) represent a specific form tumor metastasis observed in colorectal cancer (CRC). The lack of successfully established cell lines for TDs, as well as the molecular mechanisms by which TDs occur remain largely unknown. Here, we established paired CRC organoids, including a human primary cancer organoid and its TD organoid, from a 46-year-old male patient with CRC. Further analysis revealed that, compared with primary tumor-derived cells, TD-derived cells exhibited enhanced proliferative, invasive and metastatic capabilities, and increased expression of stemness-related proteins. Furthermore, the present findings also demonstrated that TD-derived cells were more resistant to oxaliplatin or 5-FU. Transcriptomic profiling and qPCR revealed that TD-derived cells exhibited more alterations in fatty acid metabolism signaling and enhanced lipid synthesis ability compared to primary tumor-derived cells. Inhibition of lipid synthesis markedly decreased resistance to oxaliplatin in TD-derived cells. Taken together, the paired organoids established using CRC primary tumor and its TD specimens will provide valuable tools to study tumorigenicity, metastasis and chemoresistance in CRC. Notably, these models will provide novel insights to study tumor heterogeneity and lipid metabolism in CRC.
Keywords: Colorectal cancer; Drug resistance; Organoid; Primary tumor; TD.
© 2024. The Author(s).
Conflict of interest statement
All authors declare no competing interests
Figures

References
-
- Siegel RL, et al. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73(3):233–54. - PubMed
-
- Bhattacharya R, et al. An insight into metastasis: random or evolving paradigms? Pathol Res Pract. 2018;214(8):1064–73. - PubMed
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Dekker E, et al. Colorectal cancer. Lancet. 2019;394(10207):1467–80. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

