DNA-directed termination of mammalian RNA polymerase II
- PMID: 39496457
- PMCID: PMC11610936
- DOI: 10.1101/gad.351978.124
DNA-directed termination of mammalian RNA polymerase II
Abstract
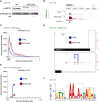
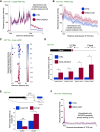
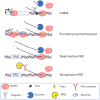
The best-studied mechanism of eukaryotic RNA polymerase II (RNAPII) transcriptional termination involves polyadenylation site-directed cleavage of the nascent RNA. The RNAPII-associated cleavage product is then degraded by XRN2, dislodging RNAPII from the DNA template. In contrast, prokaryotic RNAP and eukaryotic RNAPIII often terminate directly at T-tracts in the coding DNA strand. Here, we demonstrate a similar and omnipresent capability for mammalian RNAPII. Importantly, this termination mechanism does not require upstream RNA cleavage. Accordingly, T-tract-dependent termination can take place when XRN2 cannot be engaged. We show that T-tracts can terminate snRNA transcription independently of RNA cleavage by the Integrator complex. Importantly, we found genome-wide termination at T-tracts in promoter-proximal regions but not within protein-coding gene bodies. XRN2-dependent termination dominates downstream from protein-coding genes, but the T-tract process is sometimes used. Overall, we demonstrate global DNA-directed attrition of RNAPII transcription, suggesting that RNAPs retain the potential to terminate over T-rich sequences throughout evolution.
Keywords: Integrator; RNA polymerase II; Xrn2; exosome; histone; snRNA; transcription termination.
© 2024 Davidson et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Albrecht TR, Shevtsov SP, Wu Y, Mascibroda LG, Peart NJ, Huang KL, Sawyer IA, Tong L, Dundr M, Wagner EJ. 2018. Integrator subunit 4 is a “Symplekin-like” scaffold that associates with INTS9/11 to form the Integrator cleavage module. Nucleic Acids Res 46: 4241–4255. 10.1093/nar/gky100 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
