The Icelandic Mutation (APP-A673T) Is Protective against Amyloid Pathology In Vivo
- PMID: 39496485
- PMCID: PMC11580785
- DOI: 10.1523/JNEUROSCI.0223-24.2024
The Icelandic Mutation (APP-A673T) Is Protective against Amyloid Pathology In Vivo
Abstract
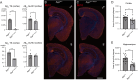
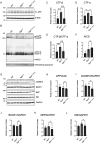
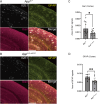
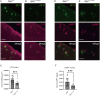
A previous epidemiological study in Northern Europe showed that the A673T mutation (Icelandic mutation) in the amyloid precursor protein gene (APP) can protect against Alzheimer's disease (AD). While the effect of the A673T mutation on APP processing has been investigated primarily in vitro, its in vivo impact has not been evaluated. This is mainly because most existing AD mouse models carry the Swedish mutation. The Swedish and Icelandic mutations are both located near the β-cleavage site, and each mutation is presumed to have the opposite effect on β-cleavage. Therefore, in the AD mouse models with the Swedish mutation, its effects could compete with the effects of the Icelandic mutation. Here, we introduced the A673T mutation into App knock-in mice devoid of the Swedish mutation (AppG-F mice) to avoid potential deleterious effects of the Swedish mutation and generated AppG-F-A673T mice. APP-A673T significantly downregulated β-cleavage and attenuated the production of Aβ and amyloid pathology in the brains of these animals. The Icelandic mutation also reduced neuroinflammation and neuritic alterations. Both sexes were studied. This is the first successful demonstration of the protective effect of the Icelandic mutation on amyloid pathology in vivo. Our findings indicate that specific inhibition of the APP-BACE1 interaction could be a promising therapeutic approach. Alternatively, the introduction of the disease-protective mutation such as APP-A673T using in vivo genome editing technology could be a novel treatment for individuals at high risk for AD, such as familial AD gene mutation carriers and APOE ε4 carriers.
Keywords: Alzheimer’s disease; App knock-in mice; Icelandic mutation; amyloid pathology; amyloid precursor protein; protective mutation.
Copyright © 2024 the authors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
