Multi-bioinformatics revealed potential biomarkers and repurposed drugs for gastric adenocarcinoma-related gastric intestinal metaplasia
- PMID: 39496635
- PMCID: PMC11535201
- DOI: 10.1038/s41540-024-00455-0
Multi-bioinformatics revealed potential biomarkers and repurposed drugs for gastric adenocarcinoma-related gastric intestinal metaplasia
Abstract
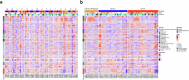
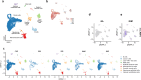
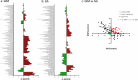
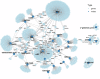
Biomarkers associated with the progression from gastric intestinal metaplasia (GIM) to gastric adenocarcinoma (GA), i.e., GA-related GIM, could provide valuable insights into identifying patients with increased risk for GA. The aim of this study was to utilize multi-bioinformatics to reveal potential biomarkers for the GA-related GIM and predict potential drug repurposing for GA prevention in patients. The multi-bioinformatics included gene expression matrix (GEM) by microarray gene expression (MGE), ScType (a fully automated and ultra-fast cell-type identification based solely on a given scRNA-seq data), Ingenuity Pathway Analysis, PageRank centrality, GO and MSigDB enrichments, Cytoscape, Human Protein Atlas and molecular docking analysis in combination with immunohistochemistry. To identify GA-related GIM, paired surgical biopsies were collected from 16 GIM-GA patients who underwent gastrectomy, yielding 64 samples (4 biopsies per stomach x 16 patients) for MGE. Co-analysis was performed by including scRNAseq and immunohistochemistry datasets of endoscopic biopsies of 37 patients. The results of the present study showed potential biomarkers for GA-related GIM, including GEM of individual patients, individual genes (such as RBP2 and CD44), signaling pathways, network of molecules, and network of signaling pathways with key topological nodes. Accordingly, potential treatment targets with repurposed drugs were identified including epidermal growth factor receptor, proto-oncogene tyrosine-protein kinase Src, paxillin, transcription factor Jun, breast cancer type 1 susceptibility protein, cellular tumor antigen p53, mouse double minute 2, and CD44.
© 2024. The Author(s).
Conflict of interest statement
Parts of the results were included in an MSc thesis by S. Geithus (
Figures

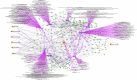

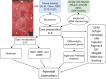
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

