The evolutionary landscape of prokaryotic chromosome/plasmid balance
- PMID: 39496780
- PMCID: PMC11535066
- DOI: 10.1038/s42003-024-07167-5
The evolutionary landscape of prokaryotic chromosome/plasmid balance
Abstract
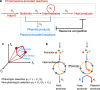
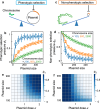
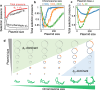
The balance between chromosomal and plasmid DNAs determines the genomic plasticity of prokaryotes. Natural selections, acting on the level of organisms or plasmids, shape the abundances of plasmid DNAs in prokaryotic genomes. Despite the importance of plasmids in health and engineering, there have been rare systematic attempts to quantitatively model and predict the determinants underlying the strength of different selection forces. Here, we develop a metabolic flux model that describes the intracellular resource competition between chromosomal and plasmid-encoded reactions. By coarse graining, this model predicts a landscape of natural selections on chromosome/plasmid balance, which is featured by the tradeoff between phenotypic and non-phenotypic selection pressures. This landscape is further validated by the observed pattern of plasmid distributions in the vast collection of prokaryotic genomes retrieved from the NCBI database. Our results establish a universal paradigm to understand the prokaryotic chromosome/plasmid interplay and provide insights into the evolutionary origin of plasmid diversity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources

