Molecular characterization and computational analysis of a highly specific L-glutaminase from a marine bacterium Bacillus australimaris NIOT30
- PMID: 39496784
- PMCID: PMC11535052
- DOI: 10.1038/s41598-024-77959-6
Molecular characterization and computational analysis of a highly specific L-glutaminase from a marine bacterium Bacillus australimaris NIOT30
Abstract
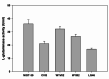
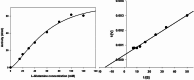
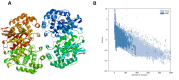
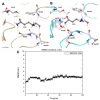
An alkaline active L-glutaminase (BALG) producing bacterium was screened and identified from seamount sediment samples of the Arabian Sea. The isolate was confirmed to be Bacillus australimaris NIOT30 based on morphological characteristics and 16 S rRNA gene sequencing. The glutaminase gene, balg was PCR amplified, cloned and expressed in E. coli BL21 (DE3) host. The molecular weight of purified BALG was estimated to be 36 kDa and the enzyme showed a specific activity of 507 ± 27 Umg-1 against L-glutamine under optimal assay conditions of pH 7.0 and temperature at 37 °C for 15 min. The enzyme showed maximum activity at pH 7 and retained 95% activity at pH 10. BALG retained a relative activity of about 82% and 45% at 45 °C and 60 °C respectively. The kinetic parameters of BALG, Km and Kcat/Km were determined to be of 210 ± 11 mM and 4.4 × 102 M s-1 respectively. Homology modeling and substrate ligand interaction studies revealed the stability of the enzyme-substrate complex. The present study highlights the characterization of a highly active L-glutaminase from B. australimaris NIOT30. Further, mutational analyses of ligand binding residues would show insights into the affinity of L-Glutaminase.
Keywords: Bacillus australimaris; Characterization; L-Glutaminase; Sediment; Specificity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
A potential type-II L-asparaginase from marine isolate Bacillus australimaris NJB19: Statistical optimization, in silico analysis and structural modeling.Int J Biol Macromol. 2021 Mar 31;174:527-539. doi: 10.1016/j.ijbiomac.2021.01.130. Epub 2021 Jan 25. Int J Biol Macromol. 2021. PMID: 33508362
-
Efficient expression and purification of recombinant glutaminase from Bacillus licheniformis (GlsA) in Escherichia coli.Protein Expr Purif. 2012 May;83(1):52-8. doi: 10.1016/j.pep.2012.03.001. Epub 2012 Mar 9. Protein Expr Purif. 2012. PMID: 22433447
-
Protein-glutaminase from Chryseobacterium proteolyticum, an enzyme that deamidates glutaminyl residues in proteins. Purification, characterization and gene cloning.Eur J Biochem. 2001 Mar;268(5):1410-21. doi: 10.1046/j.1432-1327.2001.02019.x. Eur J Biochem. 2001. PMID: 11231294
-
Exploring Escherichia coli EGY Type II L-Asparaginase Variant of Unique Glutaminase Activity: Cloning, Expression, Biochemical Characterization, and Molecular Docking Analysis.Curr Microbiol. 2025 May 25;82(7):306. doi: 10.1007/s00284-025-04233-x. Curr Microbiol. 2025. PMID: 40413673
-
Characterization of two glutaminases from the filamentous cyanobacterium Anabaena sp. PCC 7120.FEMS Microbiol Lett. 2008 Dec;289(2):241-9. doi: 10.1111/j.1574-6968.2008.01395.x. FEMS Microbiol Lett. 2008. PMID: 19054111
References
-
- Elshafei, A. & Purification Kinetic Properties and Antitumor Activity of L-Glutaminase from Penicillium brevicompactum NRC 829. Br. Microbiol. Res. J..4, 97–115 (2014).
-
- McCauley, R., Kong, S. E., Heel, K. & Hall, J. C. The role of glutaminase in the small intestine. Int. J. Biochem. Cell. Biol.31, 405–413 (1999). - PubMed
-
- Baskaran, R., Bandikari, R., Zuo, W., Qian, J. & Liu, Z. Enhanced thermostability of halo-tolerant glutaminase from Bacillus licheniformis ATCC 14580 by immobilization onto nano magnetic cellulose sheet and its application in production of glutamic acid. Int. J. Biol. Macromol.119, 1256–1263 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

