The ZmCPK39-ZmDi19-ZmPR10 immune module regulates quantitative resistance to multiple foliar diseases in maize
- PMID: 39496881
- PMCID: PMC11631770
- DOI: 10.1038/s41588-024-01968-4
The ZmCPK39-ZmDi19-ZmPR10 immune module regulates quantitative resistance to multiple foliar diseases in maize
Erratum in
-
Author Correction: The ZmCPK39-ZmDi19-ZmPR10 immune module regulates quantitative resistance to multiple foliar diseases in maize.Nat Genet. 2025 Feb;57(2):480. doi: 10.1038/s41588-025-02091-8. Nat Genet. 2025. PMID: 39838095 Free PMC article. No abstract available.
Abstract
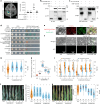
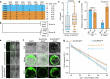
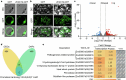
Gray leaf spot, northern leaf blight and southern leaf blight are three of the most destructive foliar diseases affecting maize (Zea mays L.). Here we identified a gene, ZmCPK39, that encodes a calcium-dependent protein kinase and negatively regulates quantitative resistance to these three diseases. The ZmCPK39 allele in the resistant line displayed significantly lower pathogen-induced gene expression than that in the susceptible line. A marked decrease in ZmCPK39 abundance mitigated the phosphorylation and degradation of the transcription factor ZmDi19. This led to elevated expression of ZmPR10, a gene known to encode an antimicrobial protein, thereby enhancing maize resistance to foliar diseases. Moreover, the F1 hybrid with reduced ZmCPK39 expression favored disease resistance, thereby increasing yield. Hence, the discovery of the ZmCPK39-ZmDi19-ZmPR10 immune module provides insight into the mechanisms underlying broad-spectrum quantitative disease resistance and also offers a new avenue for the genetic control of maize foliar diseases.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures














References
-
- Chang, H. S. & Fan, K. C. Comparative studies on some biology and pathology of corn and broom corn isolates of Exserohilum turcicum (Pass) Leonard & Suggs. Bot. Bull. Acad. Sin.27, 209–218 (1986).
-
- Ward, J. M. J., Laing, M. D. & Nowell, D. C. Chemical control of maize grey leaf spot. Crop Prot.16, 265–271 (1997). - DOI
-
- Juliatti, F. C., Pedrosa, M. G., Silva, H. D. & da Silva, J. V. C. Genetic mapping for resistance to gray leaf spot in maize. Euphytica169, 227–238 (2009). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

