Novel coenzyme Q6 genetic variant increases susceptibility to pneumococcal disease
- PMID: 39496954
- PMCID: PMC11908386
- DOI: 10.1038/s41590-024-01998-4
Novel coenzyme Q6 genetic variant increases susceptibility to pneumococcal disease
Abstract
Acute lower respiratory tract infection (ALRI) remains a major worldwide cause of childhood mortality, compelling innovation in prevention and treatment. Children in Papua New Guinea (PNG) experience profound morbidity from ALRI caused by Streptococcus pneumoniae. As a result of evolutionary divergence, the human PNG population exhibits profound genetic variation and diversity. To address unmet health needs of children in PNG, we tested whether genetic variants increased ALRI morbidity. Whole-exome sequencing of a pilot child cohort identified homozygosity for a novel single-nucleotide variant (SNV) in coenzyme Q6 (COQ6) in cases with ALRI. COQ6 encodes a mitochondrial enzyme essential for biosynthesis of ubiquinone, an electron acceptor in the electron transport chain. A significant association of SNV homozygosity with ALRI was replicated in an independent ALRI cohort (P = 0.036). Mice homozygous for homologous mouse variant Coq6 exhibited increased mortality after pneumococcal lung infection, confirming causality. Bone marrow chimeric mice further revealed that expression of variant Coq6 in recipient (that is, nonhematopoietic) tissues conferred increased mortality. Variant Coq6 maintained ubiquinone biosynthesis, while accelerating metabolic remodeling after pneumococcal challenge. Identification of this COQ6 variant provides a genetic basis for increased pneumonia susceptibility in PNG and establishes a previously unrecognized role for the enzyme COQ6 in regulating inflammatory-mediated metabolic remodeling.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: T.E.D. had no financial conflicts of interest during the years in which he contributed to the project (2018 and earlier). He is currently employed by Mission Bio, Inc. and serves as a scientific advisor for RhoDx, Inc. The other authors declare no competing interests.
Figures





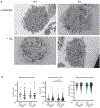
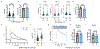
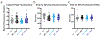



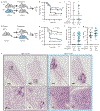




References
References (main text)
References (Methods only)
MeSH terms
Substances
Grants and funding
- R01 HL177453/HL/NHLBI NIH HHS/United States
- PD-II-2018-742/Children's Discovery Institute (CDI)
- R01 AI139540/AI/NIAID NIH HHS/United States
- R01 AI166793/AI/NIAID NIH HHS/United States
- F32 DK136180/DK/NIDDK NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- R01 HL141086/HL/NHLBI NIH HHS/United States
- P30 DK020579/DK/NIDDK NIH HHS/United States
- PD-II-2013-295/Children's Discovery Institute (CDI)
- R21 AI142723/AI/NIAID NIH HHS/United States
- PD-II-2014-366/Children's Discovery Institute (CDI)
- P30 CA091842/CA/NCI NIH HHS/United States
- T32 AI007172/AI/NIAID NIH HHS/United States
- R01 AI036478/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

