The molecular consequences of FOXF1 missense mutations associated with alveolar capillary dysplasia with misalignment of pulmonary veins
- PMID: 39497128
- PMCID: PMC11536904
- DOI: 10.1186/s12929-024-01088-5
The molecular consequences of FOXF1 missense mutations associated with alveolar capillary dysplasia with misalignment of pulmonary veins
Abstract
Background: Alveolar capillary dysplasia with misalignment of pulmonary veins (ACD/MPV) is a fatal congenital lung disorder strongly associated with genomic alterations in the Forkhead box F1 (FOXF1) gene and its regulatory region. However, little is known about how FOXF1 genomic alterations cause ACD/MPV and what molecular mechanisms are affected by these mutations. Therefore, the effect of ACD/MPV patient-specific mutations in the FOXF1 gene on the molecular function of FOXF1 was studied.
Methods: Epitope-tagged FOXF1 constructs containing one of the ACD/MPV-associated mutations were expressed in mammalian cell lines to study the effect of FOXF1 mutations on protein function. EMSA binding assays and luciferase assays were performed to study the effect on target gene binding and activation. Immunoprecipitation followed by SDS‒PAGE and western blotting were used to study protein‒protein interactions. Protein phosphorylation was studied using phos-tag western blotting.
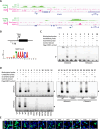
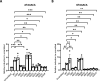
Results: An overview of the localization of ACD/MPV-associated FOXF1 mutations revealed that the G91-S101 region was frequently mutated. A three-dimensional model of the forkhead DNA-binding domain of FOXF1 showed that the G91-S101 region consists of an α-helix and is predicted to be important for DNA binding. We showed that FOXF1 missense mutations in this region differentially affect the DNA binding of the FOXF1 protein and influence the transcriptional regulation of target genes depending on the location of the mutation. Furthermore, we showed that some of these mutations can affect the FOXF1 protein at the posttranscriptional level, as shown by altered phosphorylation by MST1 and MST2 kinases.
Conclusion: Missense mutations in the coding region of the FOXF1 gene alter the molecular function of the FOXF1 protein at multiple levels, such as phosphorylation, DNA binding and target gene activation. These results indicate that FOXF1 molecular pathways may be differentially affected in ACD/MPV patients carrying missense mutations in the DNA-binding domain and may explain the phenotypic heterogeneity of ACD/MPV.
Keywords: ACD/MPV; Alveolar capillary dysplasia with misalignment of the pulmonary veins; DNA-binding domain; FOXF1; Missense mutations; Phosphorylation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures



References
-
- Steiner LA, Getman M, Schiralli Lester GM, Iqbal MA, Katzman P, Szafranski P, et al. Disruption of normal patterns of FOXF1 expression in a lethal disorder of lung development. J Med Genet. 2020;57(5):296–300. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

