Molecular Dynamics Investigation of the Influenza Hemagglutinin Conformational Changes in Acidic pH
- PMID: 39497238
- PMCID: PMC11571222
- DOI: 10.1021/acs.jpcb.4c04607
Molecular Dynamics Investigation of the Influenza Hemagglutinin Conformational Changes in Acidic pH
Abstract
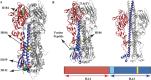
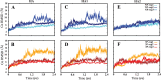
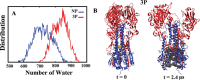
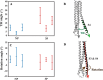
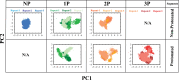
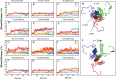
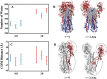
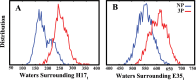
The surface protein hemagglutinin (HA) of the influenza virus plays a pivotal role in facilitating viral infection by binding to sialic acid receptors on host cells. Its conformational state is pH-sensitive, impacting its receptor-binding ability and evasion of the host immune response. In this study, we conducted extensive equilibrium microsecond-level all-atom molecular dynamics (MD) simulations of the HA protein to explore the influence of low pH on its conformational dynamics. Specifically, we investigated the impact of protonation on conserved histidine residues (H1062) located in the hinge region of HA2. Our analysis encompassed comparisons between nonprotonated (NP), partially protonated (1P, 2P), and fully protonated (3P) conditions. Our findings reveal substantial pH-dependent conformational alterations in the HA protein, affecting its receptor-binding capability and immune evasion potential. Notably, the nonprotonated form exhibits greater stability compared to protonated states. Conformational shifts in the central helices of HA2 involve outward movement, counterclockwise rotation of protonated helices, and fusion peptide release in protonated systems. Disruption of hydrogen bonds between the fusion peptide and central helices of HA2 drives this release. Moreover, HA1 separation is more likely in the fully protonated system (3P) compared to nonprotonated systems (NP), underscoring the influence of protonation. These insights shed light on influenza virus infection mechanisms and may inform the development of novel antiviral drugs targeting HA protein and pH-responsive drug delivery systems for influenza.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Update of
-
Molecular dynamics investigation of the influenza hemagglutinin conformational changes in acidic pH.bioRxiv [Preprint]. 2024 Jul 10:2024.07.07.602399. doi: 10.1101/2024.07.07.602399. bioRxiv. 2024. Update in: J Phys Chem B. 2024 Nov 14;128(45):11151-11163. doi: 10.1021/acs.jpcb.4c04607. PMID: 39026831 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

