Single-Cell Transcriptomics Reveals STAT1 Drives LHPP Downregulation in Esophageal Squamous Cell Carcinoma Progression
- PMID: 39497541
- PMCID: PMC11536641
- DOI: 10.1177/15330338241293485
Single-Cell Transcriptomics Reveals STAT1 Drives LHPP Downregulation in Esophageal Squamous Cell Carcinoma Progression
Abstract
Background: Esophageal Squamous Cell Carcinoma (ESCC) poses a significant health challenge in China due to its high incidence and mortality. Single-cell transcriptomics has transformed the approach to cancer research, allowing detailed analysis of cellular and molecular heterogeneity. The interaction between the transcription factor STAT1 and the tumor suppressor LHPP is crucial, potentially influencing cancer progression and therapeutic outcomes.
Objective: This study aims to explore the molecular mechanisms and cellular dynamics underlying ESCC, with a focus on the role of transcription factors, particularly STAT1, and its regulatory relationship with LHPP, in the pathogenesis of ESCC.
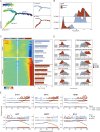
Methods: Utilizing single-cell transcriptomics data sourced from the public database GEO (GSE160269), we identified major cell types and their transcriptomic changes in ESCC patients. Differential gene expression profiles were examined to understand the dynamics of the tumor microenvironment (TME). A cohort of 21 ESCC patients was recruited to validate the findings. Furthermore, in ESCC cell lines, we validated the transcriptional regulatory relationship between STAT1 and LHPP.
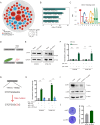
Results: Our analysis identified six major cell types within the ESCC microenvironment, revealing significant changes in cellular composition and gene expression profiles associated with tumorigenesis. Notably, a novel association between STAT1's regulatory role over LHPP was observed, suggesting a complex regulatory loop in ESCC pathogenesis. Elevated STAT1 and reduced LHPP expression were confirmed in patient samples with STAT1 negatively regulates LHPP expression at the promoter level; when the promoter binding motif regions were mutated, the transcriptional repression ability on LHPP was weakened.
Conclusion: The study highlights STAT1 as a core regulator in ESCC, directly influencing LHPP expression. The findings offer novel insights into the molecular mechanisms driving ESCC, shedding light on the cellular alterations and gene regulation dynamics within the ESCC microenvironment. This research provides a foundation for developing targeted therapeutic strategies, potentially utilizing the STAT1-LHPP axis as a focal point in ESCC treatment and prognosis.
Keywords: esophageal squamous cell carcinoma; signal transducer and activator of transcription 1; single-cell trascriptomics; tumor microenvironment.
Conflict of interest statement
Declaration of Conflicting InterestsThe authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646-674. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7-30. - PubMed
-
- Morgan E, Soerjomataram I, Rumgay H, et al. The global landscape of esophageal squamous cell carcinoma and esophageal adenocarcinoma incidence and mortality in 2020 and projections to 2040: New estimates from GLOBOCAN 2020. Gastroenterology. 2022;163(3):649-658. e2. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

