Isolation and Comprehensive Analysis of Cochlear Tissue-Derived Small Extracellular Vesicles
- PMID: 39497619
- PMCID: PMC11672270
- DOI: 10.1002/advs.202408964
Isolation and Comprehensive Analysis of Cochlear Tissue-Derived Small Extracellular Vesicles
Abstract
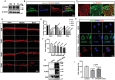
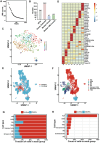
Small extracellular vesicles (sEVs) act as a critical mediator in intercellular communication. Compared to sEVs derived from in vitro sources, tissue-derived sEVs can reflect the in vivo signals released from specific tissues more accurately. Currently, studies on the role of sEVs in the cochlea have relied on studying sEVs from in vitro sources. This study evaluates three cochlear tissue digestion and cochlear tissue-derived sEV (CDsEV) isolation methods, and first proposes that the optimal approach for isolating CDsEVs using collagenase D and DNase І combined with sucrose density gradient centrifugation. Furthermore, it comprehensively investigates CDsEV contents and cell origins. Small RNA sequencing and proteomics are performed to analyze the miRNAs and proteins of CDsEVs. The miRNAs and proteins of CDsEVs are crucial for maintaining normal auditory function. Among them, FGFR1 in CDsEVs may mediate the survival of cochlear hair cells via sEVs. Finally, the joint analysis of single CDsEV sequencing and single-cell RNA sequencing data is utilized to trace cellular origins of CDsEVs. The results show that different types of cochlear cells secrete different amounts of CDsEVs, with Kölliker's organ cells and supporting cells secrete the most. The findings are expected to enhance the understanding of CDsEVs in the cochlea.
Keywords: FGFR1; cochlea; hair cells; miRNA; small extracellular vesicle; supporting cells.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- a) Théry C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin‐Smith G. K., Ayre D. C., Bach J. M., Bachurski D., Baharvand H., Balaj L., Baldacchino S., Bauer N. N., Baxter A. A., Bebawy M., Beckham C., Bedina Zavec A., Benmoussa A., Berardi A. C., Bergese P., Bielska E., Blenkiron C., Bobis‐Wozowicz S., Boilard E., Boireau W., Bongiovanni A., et al., J. Extracell. Vesicles 2018, 7, 1535750; - PMC - PubMed
- b) Welsh J. A., Goberdhan D. C. I., O'Driscoll L., Buzas E. I., Blenkiron C., Bussolati B., Cai H., Di Vizio D., Driedonks T. A. P., Erdbrügger U., Falcon‐Perez J. M., Fu Q. L., Hill A. F., Lenassi M., Lim S. K., Mahoney M. G., Mohanty S., Möller A., Nieuwland R., Ochiya T., Sahoo S., Torrecilhas A. C., Zheng L., Zijlstra A., Abuelreich S., Bagabas R., Bergese P., Bridges E. M., Brucale M., Burger D., et al., J. Extracell. Vesicles 2024, 13, e12404. - PubMed
-
- a) Zuo R., Ye L. F., Huang Y., Song Z. Q., Wang L., Zhi H., Zhang M. Y., Li J. Y., Zhu L., Xiao W. J., Shang H. C., Zhang Y., He R. R., Chen Y., J. Nanobiotechnol. 2021, 19, 396; - PMC - PubMed
- b) Xu C., Zhang Z., Liu N., Li L., Zhong H., Wang R., Shi Q., Zhang Z., Wei L., Hu B., Zhang H., Shen X., Wang Y., Liu Y., Yuan W., Nat. Commun. 2022, 13, 2467; - PMC - PubMed
- c) Liu Y., Zhang Z., Wang B., Dong Y., Zhao C., Zhao Y., Zhang L., Liu X., Guo J., Chen Y., Zhou J., Yang T., Wang Y., Liu H., Wang S., Small 2022, 18, e2107354. - PubMed
-
- a) Lee Y., Ni J., Beretov J., Wasinger V. C., Graham P., Li Y., Mol. Cancer 2023, 22, 33; - PMC - PubMed
- b) Mahmoudi K., Ezrin A., Hadjipanayis C., Mol. Aspects Med. 2015, 45, 97; - PubMed
- c) Stuendl A., Kraus T., Chatterjee M., Zapke B., Sadowski B., Moebius W., Hobert M. A., Deuschle C., Brockmann K., Maetzler W., Mollenhauer B., Schneider A., Mov. Disord. 2021, 36, 2508. - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFA1801804/National Key Research and Development Program of China
- 2022YFA0807000/National Key Research and Development Program of China
- 2021YFA1101300/National Key Research and Development Program of China
- 2021YFA1101800/National Key Research and Development Program of China
- 2020YFA0112503/National Key Research and Development Program of China
- 82171149/National Natural Science Foundation of China
- 82371166/National Natural Science Foundation of China
- 81970892/National Natural Science Foundation of China
- 82330033/National Natural Science Foundation of China
- 82030029/National Natural Science Foundation of China
- 81970882/National Natural Science Foundation of China
- 92149304/National Natural Science Foundation of China
- JCYJ20230807114700001/Shenzhen Science and Technology Program
- JCYJ20210324125608022/Shenzhen Science and Technology Program
- JCYJ20190814093401920/Shenzhen Science and Technology Program
- JCYJ20190808120405672/Shenzhen Science and Technology Program
- 2024A1515010548/Basic and Applied Basic Research Foundation of Guangdong Province
- 2021YFS0371/Science and Technology Department of Sichuan Province
- YKY-KF202201/Open Research Fund of Guangdong Academy of Medical Sciences
- BK20233002/Jiangsu Provincial Scientific Research Center of Applied Mathematics
LinkOut - more resources
Full Text Sources
Miscellaneous
