Design, synthesis, molecular docking, and dynamics studies of novel thiazole-Schiff base derivatives containing a fluorene moiety and the assessment of their antimicrobial and antioxidant activity
- PMID: 39497776
- PMCID: PMC11533417
- DOI: 10.1039/d4ra04197f
Design, synthesis, molecular docking, and dynamics studies of novel thiazole-Schiff base derivatives containing a fluorene moiety and the assessment of their antimicrobial and antioxidant activity
Abstract
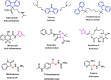
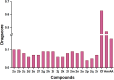
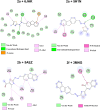
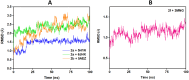
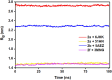
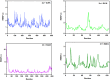
In this study, a series of eighteen fluorene-containing substituted thiazole derivatives were synthesized and characterized via spectral analyses. The proposed compounds were screened for their in vitro antimicrobial activity, and it was found that compound 2a displayed a significant zone of inhibition (20.3 ± 0.6 mm) against B. subtilis and compound 2b exhibited inhibitory activity (30.3 ± 0.6 mm) against a C. albicans fungal strain. Furthermore, antioxidant activity was evaluated for all analogues, where 2f exhibited a four-fold higher antioxidant capability (11.73 ± 1.22 μg mL-1) than the standard ascorbic acid. Oral bioavailability and toxicological parameters were considered, and most of the compounds satisfied Lipinski's rule of five and Veber's rule, except for one violation by a few derivatives. Molecular docking and molecular dynamics simulation were performed, providing more explicit ideas on the binding interaction and stability of compounds that exhibited wet lab activity. Average RMSD and RMSF values ranged between 0.5 Å and 2.5 Å, which indicated the stability of ligands inside the complex, yielding some engrossing insights.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures







References
-
- Kaboré M. Konaté I. Cissoko Y. Guindo I. Coulibaly B. Hermine M. Oumar A. A. Soumaré M. Fofana A. Zaré A. Cissé M. A. Sogoba D. Magassouba O. Issa H. H. Dao F. S. Microbiological Assessment and Antimicrobials' Use in an Infectious Diseases Department in Mali. J. Adv. Microbiol. 2021;11:8. doi: 10.4236/aim.2021.118029. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

