The terroir of Tempeh: Strong region-specific signatures in the bacterial community structures across Indonesia
- PMID: 39497934
- PMCID: PMC11533015
- DOI: 10.1016/j.crmicr.2024.100287
The terroir of Tempeh: Strong region-specific signatures in the bacterial community structures across Indonesia
Abstract
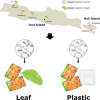
Tempeh, a soybean product from Indonesia, is created through fermentation by Rhizopus spp. and associated bacteria. Here, we aim to get an overview of the variability of the tempeh microbiota across Indonesia and disentangle influencing factors. We found high variability in bacterial abundance (103 - 109 copies g-1), richness (nASV = 40 - 175 ASVs), and diversity (H' = 0.9 - 3.5) in tempeh. The primary factor affecting this variation was the region, where the tempeh was produced. Interestingly, tempeh samples obtained from geographically close areas tended to share similar bacterial profiles, suggesting a "terroir" of tempeh. Additionally, tempeh wrapped in banana leaves had a higher abundance of enterobacteria in comparison to tempeh wrapped in plastic but also tended to have a higher total bacterial and lactobacilli abundance. Despite all variability, the tempeh core microbiome consists Lactobacillales and Enterobacteriales. This study demonstrates a high variability of bacterial diversity in traditional tempeh from local producers highlighting a strong regional influence across Indonesia.
Keywords: Amplicon sequencing; Fermented food; Lactobacilli; Microbiome; Tempeh; Terroir.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Ahnan-Winarno A.D., Cordeiro L., Winarno F.G., Gibbons J., Xiao H. Tempeh: a semicentennial review on its health benefits, fermentation, safety, processing, sustainability, and affordability. Compr. Rev. Food Sci. Food Saf. 2021;20:1717–1767. - PubMed
-
- Barus T., Suwanto A., Wahyudi A.T., Wijaya H. Role of bacteria in tempe bitter taste formation: microbiological and molecular biological analysis based on 16S rRNA gene. Microbiol. Indones. 2008;2:4.
LinkOut - more resources
Full Text Sources
Miscellaneous

