RBPWorld for exploring functions and disease associations of RNA-binding proteins across species
- PMID: 39498484
- PMCID: PMC11701580
- DOI: 10.1093/nar/gkae1028
RBPWorld for exploring functions and disease associations of RNA-binding proteins across species
Abstract
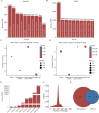
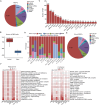
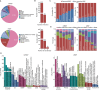
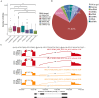
RNA-binding proteins (RBPs) play key roles in a wide range of physiological and pathological processes. To facilitate the investigation of RBP functions and disease associations, we updated the EuRBPDB and renamed it as RBPWorld (http://research.gzsys.org.cn/rbpworld/#/home). Leveraging 998 RNA-binding domains (RBDs) and 87 RNA-binding Proteome (RBPome) datasets, we successfully identified 1 393 413 RBPs from 445 species, including 3030 human RBPs (hRBPs). RBPWorld includes primary RNA targets of diverse hRBPs, as well as potential downstream regulatory pathways and alternative splicing patterns governed by various hRBPs. These insights were derived from analyses of 1515 crosslinking immunoprecipitation-seq datasets and 616 RNA-seq datasets from cells with hRBP gene knockdown or knockout. Furthermore, we systematically identified 929 RBPs with multi-functions, including acting as metabolic enzymes and transcription factors. RBPWorld includes 838 disease-associated hRBPs and 970 hRBPs that interact with 12 disease-causing RNA viruses. This provision allows users to explore the regulatory roles of hRBPs within the context of diseases. Finally, we developed an intuitive interface for RBPWorld, facilitating users easily access all the included data. We believe that RBPWorld will be a valuable resource in advancing our understanding of the biological roles of RBPs across different species.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Steinmetz E.J., Conrad N.K., Brow D.A., Corden J.L.. RNA-binding protein Nrd1 directs poly (A)-independent 3'-end formation of RNA polymerase II transcripts. Nature. 2001; 413:327–331. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

