Intracellular dynamics of ubiquitin-like 3 visualized using an inducible fluorescent timer expression system
- PMID: 39498724
- PMCID: PMC11556312
- DOI: 10.1242/bio.060345
Intracellular dynamics of ubiquitin-like 3 visualized using an inducible fluorescent timer expression system
Abstract
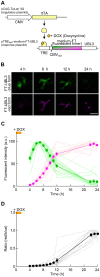
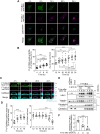
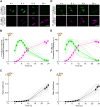
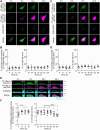
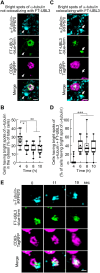
Exosomes are small extracellular vesicles (sEVs) secreted via multivesicular bodies (MVBs)/late endosomes and mediators of cell-cell communication. We previously reported a novel post-translational modification by ubiquitin-like 3 (UBL3). UBL3 is localized in MVBs and the plasma membrane and released outside as sEVs, including exosomes. Approximately 60% of proteins sorted in sEVs are affected by UBL3 and localized in various organelles, the plasma membrane, and the cytosol, suggesting that its dynamic movement in the cell before entering the MVBs. To examine the intracellular dynamics of UBL3, we constructed a sophisticated visualization system via fusing fluorescent timers that changed from blue to red form over time with UBL3 and by its expression under Tet-on regulation. Intriguingly, we found that after synthesis, UBL3 was initially distributed within the cytosol. Subsequently, UBL3 was localized to MVBs and the plasma membrane and finally showed predominant accumulation in MVBs. Furthermore, by super-resolution microscopy analysis, UBL3 was found to be associated with one of its substrates, α-tubulin, in the cytosol, and the complex was subsequently transported to MVBs. This spatiotemporal visualization system for UBL3 will form a basis for further studies to elucidate when and where UBL3 associates with its substrates/binding proteins before localization in MVBs.
Keywords: Fluorescent timers; Multivesicular bodies (MVBs); Post-translational modification (PTM); Ubiquitin-like 3 (UBL3).
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
References
-
- Ageta-Ishihara, N., Miyata, T., Ohshima, C., Watanabe, M., Sato, Y., Hamamura, Y., Higashiyama, T., Mazitschek, R., Bito, H. and Kinoshita, M. (2013). Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun. 4, 2532. 10.1038/ncomms3532 - DOI - PMC - PubMed
-
- Alberts, B., Wilson, J. H., Hunt, T., Heald, R., Johnson, A. D., Morgan, D. O., Raff, M. C., Roberts, K. and Walter, P. (2022). Molecular Biology of the Cell. W.W. Norton & Company.
Publication types
MeSH terms
Substances
Grants and funding
- JP21K07159/Japan Society for the Promotion of Science
- 22wm0525011h0002/Japan Agency for Medical Research and Development
- JPMJPR21E1/Precursory Research for Embryonic Science and Technology
- Toho University Grant for Research Initiative Program
- Astellas Foundation for Research on Metabolic Disorders
- Koyanagi-Foundation
- Chugai Foundation for Innovative Drug Discovery Science
- Research Foundation for Opto-Science and Technology
- Ministry of Education, Culture, Sports, Science, and Technology, Japan
- Intramural Research Grant (5-6) for Neurological and Psychiatric Disorders of the NCNP
- Toho University: Toho Daigaku
LinkOut - more resources
Full Text Sources

