An EED/PRC2-H19 Loop Regulates Cerebellar Development
- PMID: 39498824
- PMCID: PMC11714151
- DOI: 10.1002/advs.202403591
An EED/PRC2-H19 Loop Regulates Cerebellar Development
Abstract
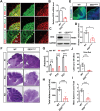
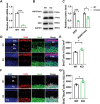
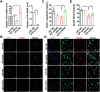
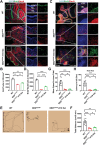
EED (embryonic ectoderm development) is a core subunit of the polycomb repressive complex 2 (PRC2), which senses the trimethylation of histone H3 lysine 27 (H3K27). However, its biological function in cerebellar development remains unknown. Here, we show that EED deletion from neural stem cells (NSCs) or cerebellar granule cell progenitors (GCPs) leads to reduced GCPs proliferation, cell death, cerebellar hypoplasia, and motor deficits in mice. Joint profiling of transcripts and ChIP-seq analysis in cerebellar granule cells reveals that EED regulates bunches of genes involved in cerebellar development. EED ablation exhibits overactivation of a developmental repressor long non-coding RNA H19. Importantly, an obvious H3K27ac enrichment is found at Ctcf, a trans-activator of H19, and H3K27me3 enrichment at the H19 imprinting control region (ICR), suggesting that EED regulates H19 in an H3K27me3-dependent manner. Intriguingly, H19 deletion reduces EED expression and the reprogramming of EED-mediated H3K27me3 profiles, resulting in increased proliferation, differentiation, and decreased apoptosis of GCPs. Finally, molecular and genetic evidence provides that increased H19 expression is responsible for cerebellar hypoplasia and motor defects in EED mutant mice. Thus, this study demonstrates that EED, H19 forms a negative feedback loop, which plays a crucial role in cerebellar morphogenesis and controls cerebellar development.
Keywords: EED, H19, Motor movement, PRC2; cerebellum.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Strick P. L., Dum R. P., Fiez J. A., Annu. Rev. Neurosci. 2009, 32, 413. - PubMed
-
- Yang Y., Yamada T., Bonni A., Handbook of the Cerebellum and Cerebellar Disorders, Springer, Berlin: 2019, 1.
-
- a) Yuen C., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R. V., Whitney J., Deflaux N., Bingham J., Wang Z., Pellecchia G., Buchanan J. A., Walker S., Marshall C. R., Uddin M., Zarrei M., Deneault E., D'Abate L., Chan A. J., Koyanagi S., Paton T., Pereira S. L., Hoang N., Engchuan W., Higginbotham E. J., Ho K., Lamoureux S., Li W., MacDonald J. R., Nalpathamkalam T., et al., Nat. Neurosci. 2017, 20, 602; - PMC - PubMed
- b) Ronan J. L., Wu W., Crabtree G. R., Nat. Rev. Genet. 2013, 14, 347; - PMC - PubMed
- c) Weiss K., Terhal P. A., Cohen L., Bruccoleri M., Irving M., Martinez A. F., Rosenfeld J. A., Machol K., Yang Y., Liu P., Walkiewicz M., Beuten J., Gomez‐Ospina N., Haude K., Fong C. T., Enns G. M., Bernstein J. A., Fan J., Gotway G., Ghorbani M., Study D. D. D., van Gassen K., Monroe G. R., van Haaften G., Basel‐Vanagaite L., Yang X. J., Campeau P. M., Muenke M., Am. J. Hum. Genet. 2016, 99, 934. - PMC - PubMed
-
- Hirabayashi Y., Suzki N., Tsuboi M., Endo T. A., Toyoda T., Shinga J., Koseki H., Vidal M., Gotoh Y., Neuron 2009, 63, 600. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
