Evaluating methods for identifying and quantifying Streptococcus pneumoniae co-colonization using next-generation sequencing data
- PMID: 39499074
- PMCID: PMC11619295
- DOI: 10.1128/spectrum.03643-23
Evaluating methods for identifying and quantifying Streptococcus pneumoniae co-colonization using next-generation sequencing data
Abstract
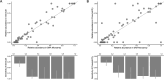
Detection of multiple pneumococcal serotype carriage can enhance monitoring of pneumococcal vaccine impact, particularly among high-burden childhood populations. We assessed methods for identifying co-carriage of pneumococcal serotypes from whole-genome sequences. Twenty-four nasopharyngeal samples were collected during community carriage surveillance from healthy children in Blantyre, Malawi, which were then serotyped by microarray. Pneumococcal DNA from culture plate sweeps were sequenced using Illumina MiSeq, and genomic serotyping was carried out using SeroCall and PneumoKITy. Their sensitivity was calculated in reference to the microarray data. Local maxima in the single-nucleotide polymorphism (SNP) density distributions were assessed for their correspondence to the relative abundance of serotypes. Across the 24 individuals, the microarray detected 77 non-unique serotypes, of which 42 occurred at high relative abundance (>10%) (per individual, median, 3; range, 1-6 serotypes). The average sequencing depth was 57X (range: 21X-88X). The sensitivity of SeroCall for identifying high-abundance serotypes was 98% (95% CI, 0.87-1.00), 20% (0.08-0.36) for low abundance (<10%), and 62% (0.50-0.72) overall. PneumoKITy's sensitivity was 86% (0.72-0.95), 20% (0.06-0.32), and 56% (0.42-0.65), respectively. Local maxima in the SNP frequency distribution were highly correlated with the relative abundance of high-abundance serotypes. Six samples were resequenced, and the pooled runs had an average fourfold increase in sequencing depth. This allowed genomic serotyping of two of the previously undetectable seven low-abundance serotypes. Genomic serotyping is highly sensitive for the detection of high-abundance serotypes in samples with co-carriage. Serotype-associated reads may be identified through SNP frequency, and increased read depth can increase sensitivity for low-abundance serotype detection.IMPORTANCEPneumococcal carriage is a prerequisite for invasive pneumococcal disease, which is a leading cause of childhood pneumonia. Multiple carriage of unique pneumococcal serotypes at a single time point is prevalent among high-burden childhood populations. This study assessed the sensitivity of different genomic serotyping methods for identifying pneumococcal serotypes during co-carriage. These methods were evaluated against the current gold standard for co-carriage detection. The results showed that genomic serotyping methods have high sensitivity for detecting high-abundance serotypes in samples with co-carriage, and increasing sequencing depth can increase sensitivity for low-abundance serotypes. These results are important for monitoring vaccine impact, which aims to reduce the prevalence of specific pneumococcal serotypes. By accurately detecting and identifying multiple pneumococcal serotypes in carrier populations, we can better evaluate the effectiveness of vaccination programs.
Keywords: Africa; Streptococcus pneumoniae; co-carriage; microarray; pneumococcus; sequencing; serotyping.
Conflict of interest statement
Jason Hinds is involved in studies at St George's, University of London, or BUGS Bioscience that are sponsored by vaccine manufacturers, including Pfizer, GlaxoSmithKline, and Sanofi Pasteur. He is also a co-founder and shareholder of BUGS Bioscience, a not-for-profit biotech company in charge of microarray results for this study out of St George's, University of London. No other authors declare competing interests.
Figures

References
-
- Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi:10.1371/journal.pgen.0020031 - DOI - PMC - PubMed
-
- Ganaie F, Saad JS, McGee L, van Tonder AJ, Bentley SD, Lo SW, Gladstone RA, Turner P, Keenan JD, Breiman RF, Nahm MH. 2020. A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral Streptococcus. mBio 11:e00937-20. doi:10.1128/mBio.00937-20 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

