Architectural dissection of adhesive bacterial cell surface appendages from a "molecular machines" viewpoint
- PMID: 39499080
- PMCID: PMC7616799
- DOI: 10.1128/jb.00290-24
Architectural dissection of adhesive bacterial cell surface appendages from a "molecular machines" viewpoint
Abstract
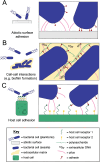
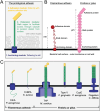
The ability of bacteria to interact with and respond to their environment is crucial to their lifestyle and survival. Bacterial cells routinely need to engage with extracellular target molecules, in locations spatially separated from their cell surface. Engagement with distant targets allows bacteria to adhere to abiotic surfaces and host cells, sense harmful or friendly molecules in their vicinity, as well as establish symbiotic interactions with neighboring cells in multicellular communities such as biofilms. Binding to extracellular molecules also facilitates transmission of information back to the originating cell, allowing the cell to respond appropriately to external stimuli, which is critical throughout the bacterial life cycle. This requirement of bacteria to bind to spatially separated targets is fulfilled by a myriad of specialized cell surface molecules, which often have an extended, filamentous arrangement. In this review, we compare and contrast such molecules from diverse bacteria, which fulfil a range of binding functions critical for the cell. Our comparison shows that even though these extended molecules have vastly different sequence, biochemical and functional characteristics, they share common architectural principles that underpin bacterial adhesion in a variety of contexts. In this light, we can consider different bacterial adhesins under one umbrella, specifically from the point of view of a modular molecular machine, with each part fulfilling a distinct architectural role. Such a treatise provides an opportunity to discover fundamental molecular principles governing surface sensing, bacterial adhesion, and biofilm formation.
Keywords: adhesins; bacterial cell surface; biofilms; fimbriae; pili.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Surface adhesins and exopolymers of selected foodborne pathogens.Microbiology (Reading). 2014 Dec;160(Pt 12):2561-2582. doi: 10.1099/mic.0.075887-0. Epub 2014 Sep 12. Microbiology (Reading). 2014. PMID: 25217529
-
Structure, Function, and Assembly of Adhesive Organelles by Uropathogenic Bacteria.Microbiol Spectr. 2015 Oct;3(5):10.1128/microbiolspec.UTI-0018-2013. doi: 10.1128/microbiolspec.UTI-0018-2013. Microbiol Spectr. 2015. PMID: 26542038 Free PMC article. Review.
-
The Mammalian Membrane Microenvironment Regulates the Sequential Attachment of Bacteria to Host Cells.mBio. 2021 Aug 31;12(4):e0139221. doi: 10.1128/mBio.01392-21. Epub 2021 Aug 3. mBio. 2021. PMID: 34340544 Free PMC article.
-
Planktonic Interference and Biofilm Alliance between Aggregation Substance and Endocarditis- and Biofilm-Associated Pili in Enterococcus faecalis.J Bacteriol. 2018 Nov 26;200(24):e00361-18. doi: 10.1128/JB.00361-18. Print 2018 Dec 15. J Bacteriol. 2018. PMID: 30249706 Free PMC article.
-
Bridging Bacteria and the Gut: Functional Aspects of Type IV Pili.Trends Microbiol. 2020 May;28(5):340-348. doi: 10.1016/j.tim.2020.02.003. Epub 2020 Mar 5. Trends Microbiol. 2020. PMID: 32298612 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

