Modelling quiescence exit of neural stem cells reveals a FOXG1-FOXO6 axis
- PMID: 39499086
- PMCID: PMC11625887
- DOI: 10.1242/dmm.052005
Modelling quiescence exit of neural stem cells reveals a FOXG1-FOXO6 axis
Abstract
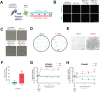
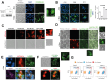
The molecular mechanisms controlling the balance of quiescence and proliferation in adult neural stem cells (NSCs) are often deregulated in brain cancers such as glioblastoma multiforme (GBM). Previously, we reported that FOXG1, a forebrain-restricted neurodevelopmental transcription factor, is frequently upregulated in glioblastoma stem cells (GSCs) and limits the effects of cytostatic pathways, in part by repression of the tumour suppressor Foxo3. Here, we show that increased FOXG1 upregulates Foxo6, a more recently discovered FOXO family member with potential oncogenic functions. Although genetic ablation of Foxo6 in proliferating NSCs had no effect on the cell cycle or entry into quiescence, we found that Foxo6-null NSCs could no longer efficiently exit quiescence following FOXG1 elevation. Increased Foxo6 resulted in the formation of large acidic vacuoles, reminiscent of Pak1-regulated macropinocytosis. Consistently, Pak1 expression was upregulated by FOXG1 overexpression and downregulated upon FOXO6 loss in proliferative NSCs. These data suggest a pro-oncogenic role for FOXO6, downstream of GBM-associated elevated FOXG1, in controlling quiescence exit, and shed light on the potential functions of this underexplored FOXO family member.
Keywords: FOXG1; FOXO6; Glioblastoma; Neural stem cell; Pak1; Quiescence.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Adusumilli, V. S., Walker, T. L., Overall, R. W., Klatt, G. M., Zeidan, S. A., Zocher, S., Kirova, D. G., Ntitsias, K., Fischer, T. J., Sykes, A. M., et al. (2021). ROS Dynamics delineate functional states of hippocampal neural stem cells and link to their activity-dependent exit from quiescence. Cell Stem Cell 28, 300-314.e6. 10.1016/j.stem.2020.10.019 - DOI - PMC - PubMed
-
- Bressan, R. B., Dewari, P. S., Kalantzaki, M., Gangoso, E., Matjusaitis, M., Garcia-Diaz, C., Blin, C., Grant, V., Bulstrode, H., Gogolok, S., et al. (2017). Efficient CRISPR/Cas9-assisted gene targeting enables rapid and precise genetic manipulation of mammalian neural stem cells. Development 144, 635-648. 10.1242/dev.140855 - DOI - PMC - PubMed
-
- Bressan, R. B., Southgate, B., Ferguson, K. M., Blin, C., Grant, V., Alfazema, N., Wills, J. C., Marques-Torrejon, M. A., Morrison, G. M., Ashmore, J., et al. (2021). Regional identity of human neural stem cells determines oncogenic responses to histone H3.3 mutants. Cell Stem Cell 28, 877-893.e9. 10.1016/j.stem.2021.01.016 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

