MoaB2, a newly identified transcription factor, binds to σA in Mycobacterium smegmatis
- PMID: 39499088
- PMCID: PMC11656743
- DOI: 10.1128/jb.00066-24
MoaB2, a newly identified transcription factor, binds to σA in Mycobacterium smegmatis
Abstract
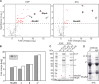
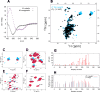
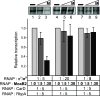
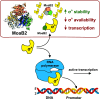
In mycobacteria, σA is the primary sigma factor. This essential protein binds to RNA polymerase (RNAP) and mediates transcription initiation of housekeeping genes. Our knowledge about this factor in mycobacteria is limited. Here, we performed an unbiased search for interacting partners of Mycobacterium smegmatis σA. The search revealed a number of proteins; prominent among them was MoaB2. The σA-MoaB2 interaction was validated and characterized by several approaches, revealing that it likely does not require RNAP and is specific, as alternative σ factors (e.g., closely related σB) do not interact with MoaB2. The structure of MoaB2 was solved by X-ray crystallography. By immunoprecipitation and nuclear magnetic resonance, the unique, unstructured N-terminal domain of σA was identified to play a role in the σA-MoaB2 interaction. Functional experiments then showed that MoaB2 inhibits σA-dependent (but not σB-dependent) transcription and may increase the stability of σA in the cell. We propose that MoaB2, by sequestering σA, has a potential to modulate gene expression. In summary, this study has uncovered a new binding partner of mycobacterial σA, paving the way for future investigation of this phenomenon.IMPORTANCEMycobacteria cause serious human diseases such as tuberculosis and leprosy. The mycobacterial transcription machinery is unique, containing transcription factors such as RbpA, CarD, and the RNA polymerase (RNAP) core-interacting small RNA Ms1. Here, we extend our knowledge of the mycobacterial transcription apparatus by identifying MoaB2 as an interacting partner of σA, the primary sigma factor, and characterize its effects on transcription and σA stability. This information expands our knowledge of interacting partners of subunits of mycobacterial RNAP, providing opportunities for future development of antimycobacterial compounds.
Keywords: MoaB2; RNA polymerase; mycobacteria; transcription; σA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

