Structure-Based Design of Covalent SARS-CoV-2 Papain-like Protease Inhibitors
- PMID: 39499574
- PMCID: PMC12182049
- DOI: 10.1021/acs.jmedchem.4c01872
Structure-Based Design of Covalent SARS-CoV-2 Papain-like Protease Inhibitors
Abstract
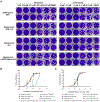
The COVID-19 pandemic is caused by SARS-CoV-2, a highly transmissible and pathogenic RNA betacoronavirus. Like other RNA viruses, SARS-CoV-2 continues to evolve with or without drug selection pressure, and many variants have emerged since the beginning of the pandemic. The papain-like protease, PLpro, is a cysteine protease that cleaves viral polyproteins as well as ubiquitin and ISG15 modifications from host proteins. Leveraging our recently discovered Val70Ub binding site in PLpro, we designed covalent PLpro inhibitors by connecting cysteine reactive warheads to the biarylphenyl PLpro inhibitors via flexible linkers. Several leads displayed potent enzymatic inhibition (IC50 = 0.1-0.3 μM) and antiviral activity (EC50 = 0.09-0.96 μM). Fumaramide inhibitors Jun13567 (15), Jun13728 (16), and Jun13714 (18) showed favorable in vivo pharmacokinetic properties with intraperitoneal injection. The X-ray crystal structure of PLpro with Jun13567 (15) validated our design strategy, revealing covalent conjugation between the catalytic Cys111 and the fumaramide warhead. The results suggest these covalent PLpro inhibitors are promising SARS-CoV-2 antiviral drug candidates.
Conflict of interest statement
Competing interests
A patent claiming the use of
Figures
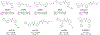








References
-
- Beigel JH; Tomashek KM; Dodd LE; Mehta AK; Zingman BS; Kalil AC; Hohmann E; Chu HY; Luetkemeyer A; Kline S; Castilla D. L. d.; Finberg RW; Dierberg K; Tapson V; Hsieh L; Patterson TF; Paredes R; Sweeney DA; Short WR; Touloumi G; Lye DC; Ohmagari N; Oh M.-d.; Ruiz-Palacios GM; Benfield T; Fätkenheuer G; Kortepeter MG; Atmar RL; Creech CB; Lundgren J; Babiker AG; Pett S; Neaton JD; Burgess TH; Bonnett T; Green M; Makowski M; Osinusi A; Nayak S; Lane HC, Remdesivir for the treatment of covid-19-final report. N. Engl. J. Med 2020, 383, 1813–1826. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

