Deciphering the RNA-binding protein network during endosomal mRNA transport
- PMID: 39499630
- PMCID: PMC11572963
- DOI: 10.1073/pnas.2404091121
Deciphering the RNA-binding protein network during endosomal mRNA transport
Abstract
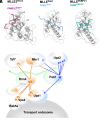
Microtubule-dependent endosomal transport is crucial for polar growth, ensuring the precise distribution of cellular cargos such as proteins and mRNAs. However, the molecular mechanism linking mRNAs to the endosomal surface remains poorly understood. Here, we present a structural analysis of the key RNA-binding protein Rrm4 from Ustilago maydis. Our findings reveal a different type of MademoiseLLE domain (MLLE) featuring a seven-helical bundle that provides a distinct binding interface. A comparative analysis with the canonical MademoiseLLE domain of the poly(A)-binding protein Pab1 disclosed unique characteristics of both domains. Deciphering the MLLE binding code enabled prediction and verification of previously unknown Rrm4 interactors containing short linear motifs. Importantly, we demonstrated that the human MLLE domains, such as those of PABPC1 and UBR5, employed a similar principle to distinguish among interaction partners. Thus, our study provides detailed mechanistic insights into how structural variations in the widely distributed MLLE domain facilitate mRNA attachment during endosomal transport.
Keywords: PAM2; RNA transport; SLiM; Ustilago maydis; endosome.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures






References
-
- Gow N. A., Hube B., Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412 (2012). - PubMed
-
- Lanver D., et al. , Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15, 409–421 (2017). - PubMed
-
- Steinberg G., The transport machinery for motility of fungal endosomes. Fungal Genet. Biol. 49, 675–676 (2012). - PubMed
MeSH terms
Substances
Grants and funding
- EXC-2048/1 - Project ID 39068111/Deutsche Forschungsgemeinschaft (DFG)
- Project-ID 267205415 - SFB 1208 to MF (project A09)/Deutsche Forschungsgemeinschaft (DFG)
- Project ID 458090666 - SFB1535/Deutsche Forschungsgemeinschaft (DFG)
- Project ID 458090666 - SFB1535/Deutsche Forschungsgemeinschaft (DFG)
- Project ID 458090666 - SFB1535/Deutsche Forschungsgemeinschaft (DFG)
LinkOut - more resources
Full Text Sources
Research Materials

