Flexibility and sensitivity in gene regulation out of equilibrium
- PMID: 39499638
- PMCID: PMC11573582
- DOI: 10.1073/pnas.2411395121
Flexibility and sensitivity in gene regulation out of equilibrium
Abstract
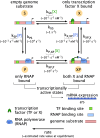
Cells adapt to environments and tune gene expression by controlling the concentrations of proteins and their kinetics in regulatory networks. In both eukaryotes and prokaryotes, experiments and theory increasingly attest that these networks can and do consume biochemical energy. How does this dissipation enable cellular behaviors forbidden in equilibrium? This open question demands quantitative models that transcend thermodynamic equilibrium. Here, we study the control of simple, ubiquitous gene regulatory networks to explore the consequences of departing equilibrium in transcription. Employing graph theory to model a set of especially common regulatory motifs, we find that dissipation unlocks nonmonotonicity and enhanced sensitivity of gene expression with respect to a transcription factor's concentration. These features allow a single transcription factor to act as both a repressor and activator at different concentrations or achieve outputs with multiple concentration regimes of locally enhanced sensitivity. We systematically dissect how energetically driving individual transitions within regulatory networks, or pairs of transitions, generates a wide range of more adjustable and sensitive phenotypic responses than in equilibrium. These results generalize to more complex regulatory scenarios, including combinatorial control by multiple transcription factors, which we relate and often find collapse to simple mathematical behaviors. Our findings quantify necessary conditions and detectable consequences of energy expenditure. These richer mathematical behaviors-feasibly accessed using biological energy budgets and rates-may empower cells to accomplish sophisticated regulation with simpler architectures than those required at equilibrium.
Keywords: biophysics; gene regulation; nonequilibrium; transcription.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





Update of
-
Flexibility and sensitivity in gene regulation out of equilibrium.bioRxiv [Preprint]. 2023 Apr 13:2023.04.11.536490. doi: 10.1101/2023.04.11.536490. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2411395121. doi: 10.1073/pnas.2411395121. PMID: 37090612 Free PMC article. Updated. Preprint.
References
-
- Wong F., Gunawardena J., Gene regulation in and out of equilibrium. Annu. Rev. Biophys. 49, 199–226 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- R35 GM118043/GM/NIGMS NIH HHS/United States
- NA/CIT | Donna and Benjamin M. Rosen Bioengineering Center, California Institute of Technology (Rosen Center, Caltech)
- R01GM139913/Foundation for the National Institutes of Health (FNIH)
- DGE-1745301/NSF | National Science Foundation Graduate Research Fellowship Program (GRFP)
- R01 GM139913/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

