The E3 ligase OsPUB33 controls rice grain size and weight by regulating the OsNAC120-BG1 module
- PMID: 39499669
- PMCID: PMC11663607
- DOI: 10.1093/plcell/koae297
The E3 ligase OsPUB33 controls rice grain size and weight by regulating the OsNAC120-BG1 module
Abstract
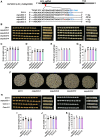
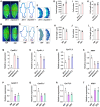
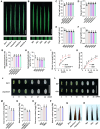
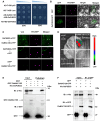
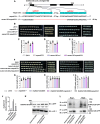
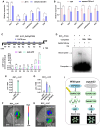
Grain size and weight are important determinants of crop yield. Although the ubiquitin pathway has been implicated in the grain development in rice (Oryza sativa), the underlying genetic and molecular mechanisms remain largely unknown. Here, we report that the plant U-box E3 ubiquitin ligase OsPUB33 interferes with the OsNAC120-BG1 module to control rice grain development. Functional loss of OsPUB33 triggers elevated photosynthetic rates and greater sugar translocation, leading to enhanced cell proliferation and accelerated grain filling. These changes cause enlarged spikelet hulls, thereby increasing final grain size and weight. OsPUB33 interacts with transcription factor OsNAC120, resulting in its ubiquitination and degradation. Unlike OsPUB33, OsNAC120 promotes grain size and weight: OsNAC120-overexpression plants harbor large and heavy grains, whereas osnac120 loss-of-function mutants produce small grains. Genetic interaction analysis supports that OsPUB33 and OsNAC120 function at least partially in a common pathway to control grain development, but have opposite functions. Additionally, OsNAC120 transcriptionally activates BIG GRAIN1 (BG1), a prominent modulator of grain size, whereas OsPUB33 impairs the OsNAC120-mediated regulation of BG1. Collectively, our findings uncover an important molecular framework for the control of grain size and weight by the OsPUB33-OsNAC120-BG1 regulatory module and provide promising targets for improving crop yield.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare no conflict of interest in this study.
Figures



References
-
- Chen W, Cheng Z, Liu L, Wang M, You X, Wang J, Zhang F, Zhou C, Zhang Z, Zhang H. Small Grain and Dwarf 2, encoding an HD-Zip II family transcription factor, regulates plant development by modulating gibberellin biosynthesis in rice. Plant Sci. 2019:288:110208. 10.1016/j.plantsci.2019.110208 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

