Community-based reconstruction and simulation of a full-scale model of the rat hippocampus CA1 region
- PMID: 39499732
- PMCID: PMC11537418
- DOI: 10.1371/journal.pbio.3002861
Community-based reconstruction and simulation of a full-scale model of the rat hippocampus CA1 region
Abstract
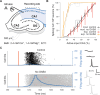
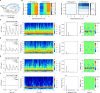
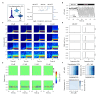
The CA1 region of the hippocampus is one of the most studied regions of the rodent brain, thought to play an important role in cognitive functions such as memory and spatial navigation. Despite a wealth of experimental data on its structure and function, it has been challenging to integrate information obtained from diverse experimental approaches. To address this challenge, we present a community-based, full-scale in silico model of the rat CA1 that integrates a broad range of experimental data, from synapse to network, including the reconstruction of its principal afferents, the Schaffer collaterals, and a model of the effects that acetylcholine has on the system. We tested and validated each model component and the final network model, and made input data, assumptions, and strategies explicit and transparent. The unique flexibility of the model allows scientists to potentially address a range of scientific questions. In this article, we describe the methods used to set up simulations to reproduce in vitro and in vivo experiments. Among several applications in the article, we focus on theta rhythm, a prominent hippocampal oscillation associated with various behavioral correlates and use our computer model to reproduce experimental findings. Finally, we make data, code, and model available through the hippocampushub.eu portal, which also provides an extensive set of analyses of the model and a user-friendly interface to facilitate adoption and usage. This community-based model represents a valuable tool for integrating diverse experimental data and provides a foundation for further research into the complex workings of the hippocampal CA1 region.
Copyright: © 2024 Romani et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Comment in
-
Gather your neurons and model together: Community times ahead.PLoS Biol. 2024 Nov 6;22(11):e3002839. doi: 10.1371/journal.pbio.3002839. eCollection 2024 Nov. PLoS Biol. 2024. PMID: 39504325 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

