Discovery of CRBN-Dependent WEE1 Molecular Glue Degraders from a Multicomponent Combinatorial Library
- PMID: 39499896
- PMCID: PMC11800961
- DOI: 10.1021/jacs.4c06127
Discovery of CRBN-Dependent WEE1 Molecular Glue Degraders from a Multicomponent Combinatorial Library
Abstract
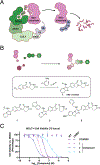
Small molecules promoting protein-protein interactions produce a range of therapeutic outcomes. Molecular glue degraders exemplify this concept due to their compact drug-like structures and ability to engage targets without reliance on existing cognate ligands. While cereblon molecular glue degraders containing glutarimide scaffolds have been approved for treatment of multiple myeloma and acute myeloid leukemia, the design of new therapeutically relevant monovalent degraders remains challenging. We report here an approach to glutarimide-containing molecular glue synthesis using multicomponent reactions as a central modular core-forming step. Screening the resulting library identified HRZ-1 derivatives that target casein kinase 1 α (CK1α) and Wee-like protein kinase (WEE1). Further medicinal chemistry efforts led to identification of selective monovalent WEE1 degraders that provide a potential starting point for the eventual development of a selective chemical degrader probe. The structure of the hit WEE1 degrader complex with CRBN-DDB1 and WEE1 provides a model of the protein-protein interface and ideas to rationalize the observed kinase selectivity. Our findings suggest that modular synthetic routes combined with in-depth structural characterization give access to selective molecular glue degraders and expansion of the CRBN-degradable proteome.
Figures




Update of
-
Discovery of CRBN-dependent WEE1 Molecular Glue Degraders from a Multicomponent Combinatorial Library.bioRxiv [Preprint]. 2024 May 5:2024.05.04.592550. doi: 10.1101/2024.05.04.592550. bioRxiv. 2024. Update in: J Am Chem Soc. 2024 Nov 20;146(46):31433-31443. doi: 10.1021/jacs.4c06127. PMID: 38746375 Free PMC article. Updated. Preprint.
References
-
- Gu S; Cui D; Chen X; Xiong X; Zhao Y. PROTACs: An Emerging Targeting Technique for Protein Degradation in Drug Discovery. BioEssays 2018, 40 (4), 1700247. - PubMed
-
- Schreiber SL The Rise of Molecular Glues. Cell 2021, 184 (1), 3–9. - PubMed
-
- Li Y-D; Ma MW; Hassan MM; Hunkeler M; Teng M; Puvar K; Lumpkin R; Sandoval B; Jin CY; Ficarro SB; Wang MY; Xu S; Groendyke BJ; Sigua LH; Tavares I; Zou C; Tsai JM; Park PMC; Yoon H; Majewski FC; Marto JA; Qi J; Nowak RP; Donovan KA; Słabicki M; Gray NS; Fischer ES; Ebert BL Template-Assisted Covalent Modification of DCAF16 Underlies Activity of BRD4Molecular Glue Degraders. bioRxiv 2023, 2023.02.14.528208.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

