Hypermethylation of CDKN2A CpG island drives resistance to PRC2 inhibitors in SWI/SNF loss-of-function tumors
- PMID: 39500892
- PMCID: PMC11538500
- DOI: 10.1038/s41419-024-07109-3
Hypermethylation of CDKN2A CpG island drives resistance to PRC2 inhibitors in SWI/SNF loss-of-function tumors
Abstract
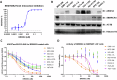
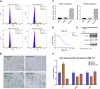
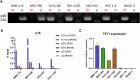
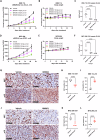
Polycomb repressive complex 2 (PRC2) catalyzes the writing of the tri-methylated histone H3 at Lys27 (H3K27me3) epigenetic marker and suppresses the expression of genes, including tumor suppressors. The function of the complex can be partially antagonized by the SWI/SNF chromatin-remodeling complex. Previous studies have suggested that PRC2 is important for the proliferation of tumors with SWI/SNF loss-of-function mutations. In the present study, we have developed an EED-directed allosteric inhibitor of PRC2 termed BR0063, which exhibits anti-proliferative properties in a subset of solid tumor cell lines harboring mutations of the SWI/SNF subunits, SMARCA4 or ARID1A. Tumor cells sensitive to BR0063 exhibited several distinct phenotypes, including cell senescence, which was mediated by the up-regulation of CDKN2A/p16. Further experiments revealed that the expression of p16 was suppressed in the BR0063-resistant cells via DNA hypermethylation in the CpG island (CGI) promoter region, rather than via PRC2 occupancy. The expression of TET1, which is required for DNA demethylation, was found to be inversely correlated with p16 CGI methylation, and this may serve as a biomarker for the prediction of resistance to PRC2 inhibitors in SWI/SNF LOF tumors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

