Plasmid-encoded phosphatase RapP enhances cell growth in non-domesticated Bacillus subtilis strains
- PMID: 39500898
- PMCID: PMC11538241
- DOI: 10.1038/s41467-024-53992-x
Plasmid-encoded phosphatase RapP enhances cell growth in non-domesticated Bacillus subtilis strains
Abstract
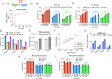
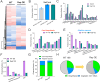
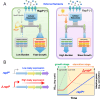
The trade-off between rapid growth and other important physiological traits (e.g., survival and adaptability) poses a fundamental challenge for microbes to achieve fitness maximization. Studies on Bacillus subtilis biology often use strains derived after a process of lab 'domestication' from an ancestral strain known as Marburg strain. The domestication process led to loss of a large plasmid (pBS32) encoding a phosphatase (RapP) that dephosphorylates the Spo0F protein and thus regulates biofilm formation and sporulation. Here, we show that plasmid pBS32, and more specifically rapP, enhance growth rates by preventing premature expression of the Spo0F-Spo0A-mediated adaptive response during exponential phase. This results in reallocation of proteome resources towards biosynthetic, growth-promoting pathways without compromising long-term fitness during stationary phase. Thus, RapP helps B. subtilis to constrain physiological trade-offs and economize cellular resources for fitness improvement.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

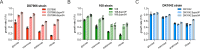



References
-
- Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science330, 1099–1102 (2010). - PubMed
-
- Reyes-Lamothe, R. & Sherratt, D. J. The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol.17, 467–478 (2019). - PubMed
-
- Kostinski, S. & Reuveni, S. Ribosome composition maximizes cellular growth rates in E. coli. Phys. Rev. Lett.125, 028103 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

