Network medicine informed multiomics integration identifies drug targets and repurposable medicines for Amyotrophic Lateral Sclerosis
- PMID: 39500920
- PMCID: PMC11538253
- DOI: 10.1038/s41540-024-00449-y
Network medicine informed multiomics integration identifies drug targets and repurposable medicines for Amyotrophic Lateral Sclerosis
Abstract
Amyotrophic Lateral Sclerosis (ALS) is a devastating, immensely complex neurodegenerative disease by lack of effective treatments. We developed a network medicine methodology via integrating human brain multi-omics data to prioritize drug targets and repurposable treatments for ALS. We leveraged non-coding ALS loci effects from genome-wide associated studies (GWAS) on human brain expression quantitative trait loci (QTL) (eQTL), protein QTL (pQTL), splicing QTL (sQTL), methylation QTL (meQTL), and histone acetylation QTL (haQTL). Using a network-based deep learning framework, we identified 105 putative ALS-associated genes enriched in known ALS pathobiological pathways. Applying network proximity analysis of predicted ALS-associated genes and drug-target networks under the human protein-protein interactome (PPI) model, we identified potential repurposable drugs (i.e., Diazoxide and Gefitinib) for ALS. Subsequent validation established preclinical evidence for top-prioritized drugs. In summary, we presented a network-based multi-omics framework to identify drug targets and repurposable treatments for ALS and other neurodegenerative disease if broadly applied.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
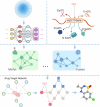
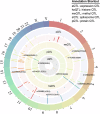

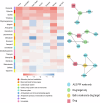
Update of
-
Network medicine informed multi-omics integration identifies drug targets and repurposable medicines for Amyotrophic Lateral Sclerosis.bioRxiv [Preprint]. 2024 Mar 30:2024.03.27.586949. doi: 10.1101/2024.03.27.586949. bioRxiv. 2024. Update in: NPJ Syst Biol Appl. 2024 Nov 5;10(1):128. doi: 10.1038/s41540-024-00449-y. PMID: 38585774 Free PMC article. Updated. Preprint.
References
-
- Mehta, P. et al. Prevalence of amyotrophic lateral sclerosis (ALS), United States, 2016. Amyotroph. Lateral Scler. Frontotemporal Degeneration.23, 220–225 (2022). - PubMed
-
- Boillée, S., Vande Velde, C. & Cleveland, D. W. ALS: A Disease of Motor Neurons and Their Nonneuronal Neighbors. Neuron52, 39–59 (2006). - PubMed
MeSH terms
Grants and funding
- R21 AG083003/AG/NIA NIH HHS/United States
- R01 AG082118/AG/NIA NIH HHS/United States
- R56 AG074001/AG/NIA NIH HHS/United States
- RF1 AG082211/AG/NIA NIH HHS/United States
- R01 AG084250/AG/NIA NIH HHS/United States
- RF1 NS133812/NS/NINDS NIH HHS/United States
- RF1NS133812/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- U01 AG073323/AG/NIA NIH HHS/United States
- U01AG073323/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01AG082118/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01AG066707/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01 AG066707/AG/NIA NIH HHS/United States
- R01AG076448/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R21AG083003/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01 AG076448/AG/NIA NIH HHS/United States
- RF1AG082211/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01AG084250/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

