Reducing competition between msd and genomic DNA improves retron editing efficiency
- PMID: 39501049
- PMCID: PMC11624263
- DOI: 10.1038/s44319-024-00311-6
Reducing competition between msd and genomic DNA improves retron editing efficiency
Abstract
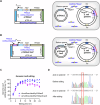
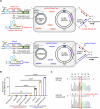
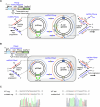
Retrons, found in bacteria and used for defense against phages, generate a unique molecule known as multicopy single-stranded DNA (msDNA). This msDNA mimics Okazaki fragments during DNA replication, making it a promising tool for targeted gene editing in prokaryotes. However, existing retron systems often exhibit suboptimal editing efficiency. Here, we identify the msd gene in Escherichia coli, which encodes the noncoding RNA template for msDNA synthesis and carries the homologous sequence of the target gene to be edited, as a critical bottleneck. Sequence homology causes the msDNA to bind to the msd gene, thereby reducing its efficiency in editing the target gene. To address this issue, we engineer a retron system that tailors msDNA to the leading strand of the plasmid containing the msd gene. This strategy minimizes msd gene editing and reduces competition with target genes, significantly increasing msDNA availability. Our optimized system achieves very high retron editing efficiency, enhancing performance and expanding the potential for in vivo techniques that rely on homologous DNA synthesis.
Keywords: Escherichia coli; DNA Replication; Gene Editing; Retron.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures



Similar articles
-
Cell-free synthesis of the branched RNA-linked msDNA from retron-Ec67 of Escherichia coli.J Biol Chem. 1992 Jul 15;267(20):13823-9. J Biol Chem. 1992. PMID: 1378431
-
Retron-Ec107 is inserted into the Escherichia coli genome by replacing a palindromic 34bp intergenic sequence.Mol Microbiol. 1992 Feb;6(3):345-54. doi: 10.1111/j.1365-2958.1992.tb01477.x. Mol Microbiol. 1992. PMID: 1372675
-
In vivo production of a stable single-stranded cDNA in Saccharomyces cerevisiae by means of a bacterial retron.Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5735-9. doi: 10.1073/pnas.89.13.5735. Proc Natl Acad Sci U S A. 1992. PMID: 1378616 Free PMC article.
-
msDNA of bacteria.Prog Nucleic Acid Res Mol Biol. 1991;40:1-24. doi: 10.1016/s0079-6603(08)60838-7. Prog Nucleic Acid Res Mol Biol. 1991. PMID: 1709507 Review.
-
Retron Library Recombineering: Next Powerful Tool for Genome Editing after CRISPR/Cas.ACS Synth Biol. 2024 Apr 19;13(4):1019-1025. doi: 10.1021/acssynbio.3c00667. Epub 2024 Mar 13. ACS Synth Biol. 2024. PMID: 38480006 Review.
Cited by
-
Genome editing of phylogenetically distinct bacteria using portable retron-mediated recombineering.bioRxiv [Preprint]. 2025 Jul 9:2025.06.16.660010. doi: 10.1101/2025.06.16.660010. bioRxiv. 2025. PMID: 40667245 Free PMC article. Preprint.
References
-
- Farzadfard F, Gharaei N, Citorik RJ, Lu TK (2021) Efficient retroelement-mediated DNA writing in bacteria. Cell Syst 12:860–872 - PubMed
MeSH terms
Substances
Grants and funding
- 31971339/MOST | National Natural Science Foundation of China (NSFC)
- 32171422/MOST | National Natural Science Foundation of China (NSFC)
- 2022YFF1000700/MOST | National Key Research and Development Program of China (NKPs)
- 2662022SKYJ004/MOE | Fundamental Research Funds for the Central Universities (Fundamental Research Fund for the Central Universities)
- GJJ211722/Jiangxi Provincial Education Department | Key Science and Technology Research Project in Jiangxi Province Department of Education
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

