Discovery of potential natural therapeutics targeting cell wall biosynthesis in multidrug-resistant Enterococcus faecalis: a computational perspective
- PMID: 39501412
- PMCID: PMC11536910
- DOI: 10.1186/s13062-024-00538-2
Discovery of potential natural therapeutics targeting cell wall biosynthesis in multidrug-resistant Enterococcus faecalis: a computational perspective
Abstract
Background: Identifying therapeutic inhibitors of crucial enzymes involved in the peptidoglycan biosynthesis pathway is pivotal for developing new treatments against multidrug-resistant Enterococcus faecalis V583. MurM, an essential enzyme in this pathway, plays a significant role in the bacterium's cell wall synthesis, making it an attractive druggable target for novel antimicrobial strategies. This study explored the potential of natural compounds as inhibitors of MurM, aiming to discover promising drug candidates that could serve as the foundation for future therapeutic development.
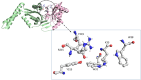
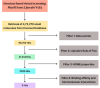
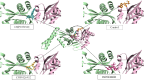
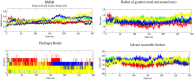
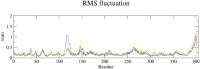
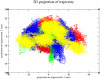
Methods: The three-dimensional structure of MurM was predicted, optimized, and its binding pocket was analyzed by comparing it with related structures. Over 4,70,000 natural compounds from the COCONUT database were subjected to virtual high-throughput screening (vHTS). The top lead candidates were selected based on their Lipinski's profile, ADME profile, toxicity profile, estimated binding free energy (ΔG) and estimated inhibition constant (Ki). Interaction pattern analysis was used to evaluate the non-covalent interactions between the inhibitors and key residues in MurM's binding pocket. Molecular dynamics simulations were performed over 300 ns to assess the structural stability and impact of these inhibitors on MurM's enzyme.
Results: Three lead compounds-CNP0056520, CNP0126952, and CNP0248480-were identified and prioritized with estimated ΔG ranging from - 9.35 to -7.9 kcal/mol. Molecular dynamics simulations revealed minimal impact on MurM's overall structure and dynamics, with the candidate inhibitors forming stable protein-ligand complexes. These interactions were supported by several non-covalent interactions between the candidate inhibitors and key residues within MurM's binding pocket.
Conclusion: These findings suggest that the identified natural product candidates could serve as promising inhibitors of MurM, potentially leading to novel therapeutics targeting cell wall biosynthesis in multidrug-resistant E. faecalis.
Keywords: Enterococcus faecalis; Molecular dynamics and antibiotic resistance; MurM; Natural compounds; Virtual screening.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
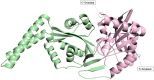


References
-
- Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, Klare I, Nallapareddy SR, Huang W, Murray BE. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003;187(3):508–12. 10.1086/367711. - PubMed
-
- Fiser A, Filipe SR, Tomasz A. Cell wall branches, penicillin resistance and the secrets of the MurM protein. Trends Microbiol. 2003;11(12):547–53. 10.1016/j.tim.2003.10.003. - PubMed
-
- Facklam RR, Carvalho MGS, Teixeira LM. History, taxonomy, biochemical characteristics and antibiotic susceptibility testing of enterococci. In: Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB, editors. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. Washington, D.C.: ASM; 2002. pp. 1–54. In.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

